Salim Arslan
Graph Saliency Maps through Spectral Convolutional Networks: Application to Sex Classification with Brain Connectivity
Jun 05, 2018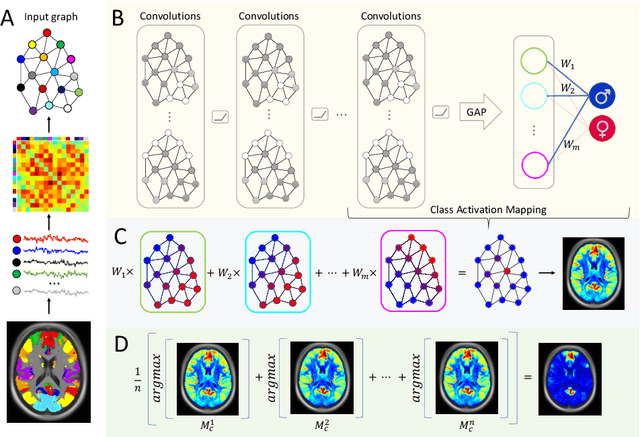
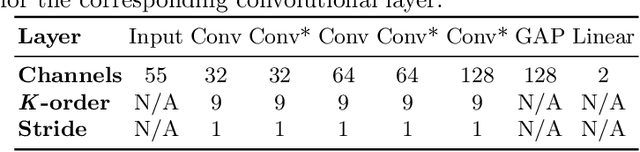
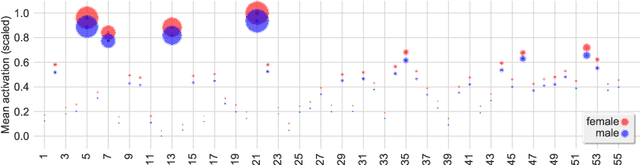
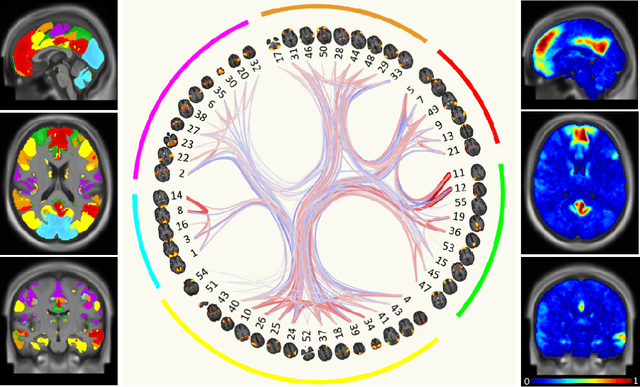
Abstract:Graph convolutional networks (GCNs) allow to apply traditional convolution operations in non-Euclidean domains, where data are commonly modelled as irregular graphs. Medical imaging and, in particular, neuroscience studies often rely on such graph representations, with brain connectivity networks being a characteristic example, while ultimately seeking the locus of phenotypic or disease-related differences in the brain. These regions of interest (ROIs) are, then, considered to be closely associated with function and/or behaviour. Driven by this, we explore GCNs for the task of ROI identification and propose a visual attribution method based on class activation mapping. By undertaking a sex classification task as proof of concept, we show that this method can be used to identify salient nodes (brain regions) without prior node labels. Based on experiments conducted on neuroimaging data of more than 5000 participants from UK Biobank, we demonstrate the robustness of the proposed method in highlighting reproducible regions across individuals. We further evaluate the neurobiological relevance of the identified regions based on evidence from large-scale UK Biobank studies.
Connectivity-Driven Parcellation Methods for the Human Cerebral Cortex
Feb 17, 2018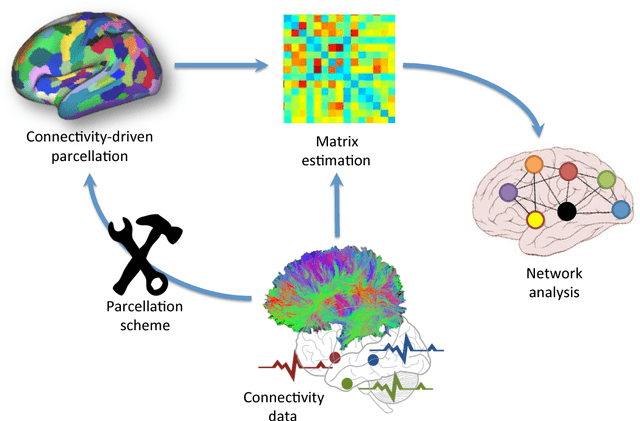
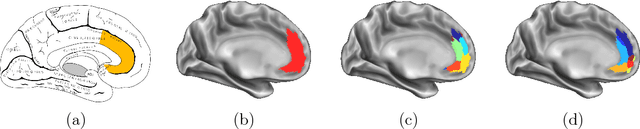
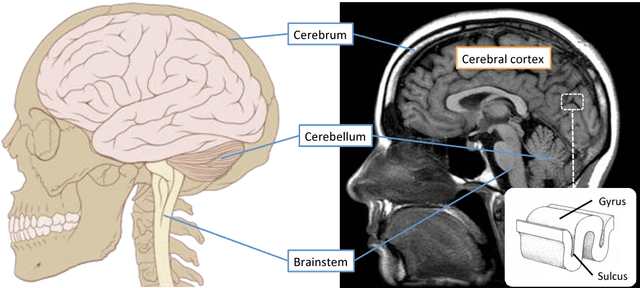
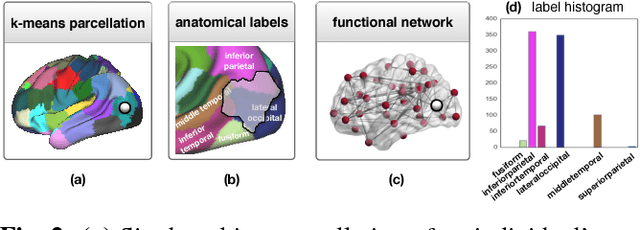
Abstract:In this thesis, we present robust and fully-automated methods for the subdivision of the entire human cerebral cortex based on connectivity information. Our contributions are four-fold: First, we propose a clustering approach to delineate a cortical parcellation that provides a reliable abstraction of the brain's functional organisation. Second, we cast the parcellation problem as a feature reduction problem and make use of manifold learning and image segmentation techniques to identify cortical regions with distinct structural connectivity patterns. Third, we present a multi-layer graphical model that combines within- and between-subject connectivity, which is then decomposed into a cortical parcellation that can represent the whole population, while accounting for the variability across subjects. Finally, we conduct a large-scale, systematic comparison of existing parcellation methods, with a focus on providing some insight into the reliability of brain parcellations in terms of reflecting the underlying connectivity, as well as, revealing their impact on network analysis. We evaluate the proposed parcellation methods on publicly available data from the Human Connectome Project and a plethora of quantitative and qualitative evaluation techniques investigated in the literature. Experiments across multiple resolutions demonstrate the accuracy of the presented methods at both subject and group levels with regards to reproducibility and fidelity to the data. The neuro-biological interpretation of the proposed parcellations is also investigated by comparing parcel boundaries with well-structured properties of the cerebral cortex. Results show the advantage of connectivity-driven parcellations over traditional approaches in terms of better fitting the underlying connectivity.
Exploring Heritability of Functional Brain Networks with Inexact Graph Matching
Mar 29, 2017
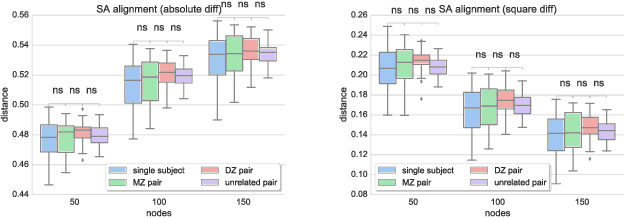
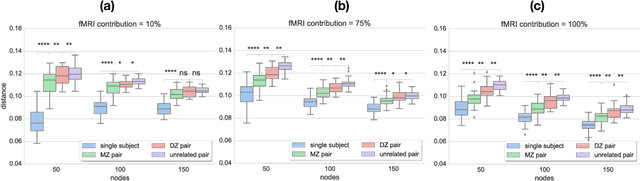
Abstract:Data-driven brain parcellations aim to provide a more accurate representation of an individual's functional connectivity, since they are able to capture individual variability that arises due to development or disease. This renders comparisons between the emerging brain connectivity networks more challenging, since correspondences between their elements are not preserved. Unveiling these correspondences is of major importance to keep track of local functional connectivity changes. We propose a novel method based on graph edit distance for the comparison of brain graphs directly in their domain, that can accurately reflect similarities between individual networks while providing the network element correspondences. This method is validated on a dataset of 116 twin subjects provided by the Human Connectome Project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge