Connectivity-Driven Parcellation Methods for the Human Cerebral Cortex
Paper and Code
Feb 17, 2018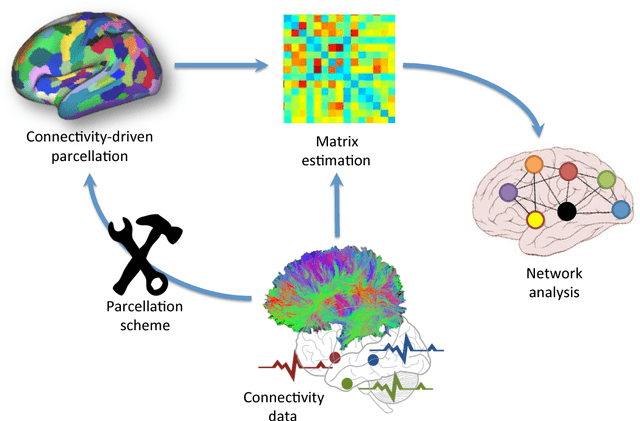
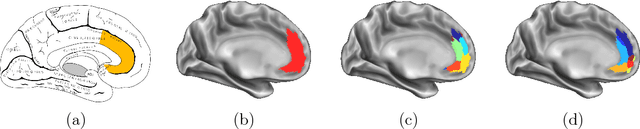
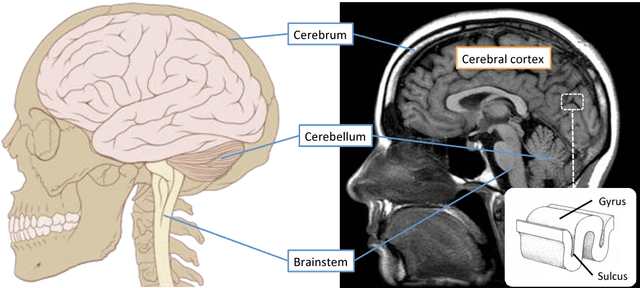
In this thesis, we present robust and fully-automated methods for the subdivision of the entire human cerebral cortex based on connectivity information. Our contributions are four-fold: First, we propose a clustering approach to delineate a cortical parcellation that provides a reliable abstraction of the brain's functional organisation. Second, we cast the parcellation problem as a feature reduction problem and make use of manifold learning and image segmentation techniques to identify cortical regions with distinct structural connectivity patterns. Third, we present a multi-layer graphical model that combines within- and between-subject connectivity, which is then decomposed into a cortical parcellation that can represent the whole population, while accounting for the variability across subjects. Finally, we conduct a large-scale, systematic comparison of existing parcellation methods, with a focus on providing some insight into the reliability of brain parcellations in terms of reflecting the underlying connectivity, as well as, revealing their impact on network analysis. We evaluate the proposed parcellation methods on publicly available data from the Human Connectome Project and a plethora of quantitative and qualitative evaluation techniques investigated in the literature. Experiments across multiple resolutions demonstrate the accuracy of the presented methods at both subject and group levels with regards to reproducibility and fidelity to the data. The neuro-biological interpretation of the proposed parcellations is also investigated by comparing parcel boundaries with well-structured properties of the cerebral cortex. Results show the advantage of connectivity-driven parcellations over traditional approaches in terms of better fitting the underlying connectivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge