Sobhan Hemati
Aggregation Schemes for Single-Vector WSI Representation Learning in Digital Pathology
Jan 29, 2025



Abstract:A crucial step to efficiently integrate Whole Slide Images (WSIs) in computational pathology is assigning a single high-quality feature vector, i.e., one embedding, to each WSI. With the existence of many pre-trained deep neural networks and the emergence of foundation models, extracting embeddings for sub-images (i.e., tiles or patches) is straightforward. However, for WSIs, given their high resolution and gigapixel nature, inputting them into existing GPUs as a single image is not feasible. As a result, WSIs are usually split into many patches. Feeding each patch to a pre-trained model, each WSI can then be represented by a set of patches, hence, a set of embeddings. Hence, in such a setup, WSI representation learning reduces to set representation learning where for each WSI we have access to a set of patch embeddings. To obtain a single embedding from a set of patch embeddings for each WSI, multiple set-based learning schemes have been proposed in the literature. In this paper, we evaluate the WSI search performance of multiple recently developed aggregation techniques (mainly set representation learning techniques) including simple average or max pooling operations, Deep Sets, Memory networks, Focal attention, Gaussian Mixture Model (GMM) Fisher Vector, and deep sparse and binary Fisher Vector on four different primary sites including bladder, breast, kidney, and Colon from TCGA. Further, we benchmark the search performance of these methods against the median of minimum distances of patch embeddings, a non-aggregating approach used for WSI retrieval.
Zero-Shot Whole Slide Image Retrieval in Histopathology Using Embeddings of Foundation Models
Sep 06, 2024Abstract:We have tested recently published foundation models for histopathology for image retrieval. We report macro average of F1 score for top-1 retrieval, majority of top-3 retrievals, and majority of top-5 retrievals. We perform zero-shot retrievals, i.e., we do not alter embeddings and we do not train any classifier. As test data, we used diagnostic slides of TCGA, The Cancer Genome Atlas, consisting of 23 organs and 117 cancer subtypes. As a search platform we used Yottixel that enabled us to perform WSI search using patches. Achieved F1 scores show low performance, e.g., for top-5 retrievals, 27% +/- 13% (Yottixel-DenseNet), 42% +/- 14% (Yottixel-UNI), 40%+/-13% (Yottixel-Virchow), and 41%+/-13% (Yottixel-GigaPath). The results for GigaPath WSI will be delayed due to the significant computational resources required for processing
Parametric Feature Transfer: One-shot Federated Learning with Foundation Models
Feb 02, 2024Abstract:In one-shot federated learning (FL), clients collaboratively train a global model in a single round of communication. Existing approaches for one-shot FL enhance communication efficiency at the expense of diminished accuracy. This paper introduces FedPFT (Federated Learning with Parametric Feature Transfer), a methodology that harnesses the transferability of foundation models to enhance both accuracy and communication efficiency in one-shot FL. The approach involves transferring per-client parametric models (specifically, Gaussian mixtures) of features extracted from foundation models. Subsequently, each parametric model is employed to generate synthetic features for training a classifier head. Experimental results on eight datasets demonstrate that FedPFT enhances the communication-accuracy frontier in both centralized and decentralized FL scenarios, as well as across diverse data-heterogeneity settings such as covariate shift and task shift, with improvements of up to 20.6%. Additionally, FedPFT adheres to the data minimization principle of FL, as clients do not send real features. We demonstrate that sending real features is vulnerable to potent reconstruction attacks. Moreover, we show that FedPFT is amenable to formal privacy guarantees via differential privacy, demonstrating favourable privacy-accuracy tradeoffs.
DFML: Decentralized Federated Mutual Learning
Feb 02, 2024Abstract:In the realm of real-world devices, centralized servers in Federated Learning (FL) present challenges including communication bottlenecks and susceptibility to a single point of failure. Additionally, contemporary devices inherently exhibit model and data heterogeneity. Existing work lacks a Decentralized FL (DFL) framework capable of accommodating such heterogeneity without imposing architectural restrictions or assuming the availability of public data. To address these issues, we propose a Decentralized Federated Mutual Learning (DFML) framework that is serverless, supports nonrestrictive heterogeneous models, and avoids reliance on public data. DFML effectively handles model and data heterogeneity through mutual learning, which distills knowledge between clients, and cyclically varying the amount of supervision and distillation signals. Extensive experimental results demonstrate consistent effectiveness of DFML in both convergence speed and global accuracy, outperforming prevalent baselines under various conditions. For example, with the CIFAR-100 dataset and 50 clients, DFML achieves a substantial increase of +17.20% and +19.95% in global accuracy under Independent and Identically Distributed (IID) and non-IID data shifts, respectively.
Analysis and Validation of Image Search Engines in Histopathology
Jan 06, 2024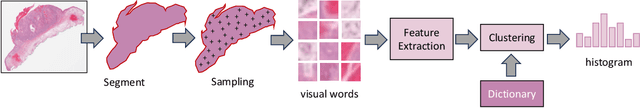
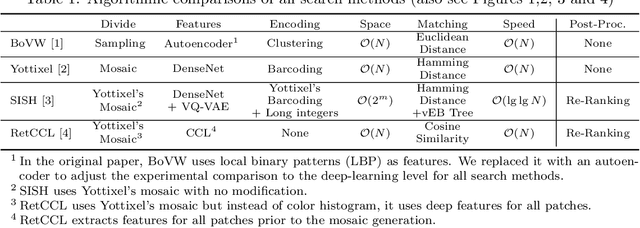
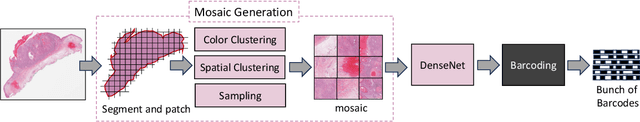
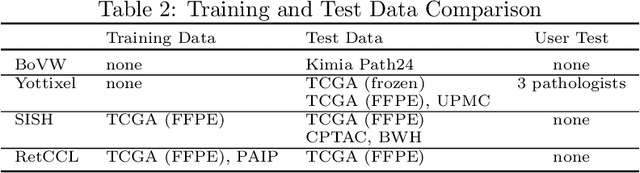
Abstract:Searching for similar images in archives of histology and histopathology images is a crucial task that may aid in patient matching for various purposes, ranging from triaging and diagnosis to prognosis and prediction. Whole slide images (WSIs) are highly detailed digital representations of tissue specimens mounted on glass slides. Matching WSI to WSI can serve as the critical method for patient matching. In this paper, we report extensive analysis and validation of four search methods bag of visual words (BoVW), Yottixel, SISH, RetCCL, and some of their potential variants. We analyze their algorithms and structures and assess their performance. For this evaluation, we utilized four internal datasets ($1269$ patients) and three public datasets ($1207$ patients), totaling more than $200,000$ patches from $38$ different classes/subtypes across five primary sites. Certain search engines, for example, BoVW, exhibit notable efficiency and speed but suffer from low accuracy. Conversely, search engines like Yottixel demonstrate efficiency and speed, providing moderately accurate results. Recent proposals, including SISH, display inefficiency and yield inconsistent outcomes, while alternatives like RetCCL prove inadequate in both accuracy and efficiency. Further research is imperative to address the dual aspects of accuracy and minimal storage requirements in histopathological image search.
Cross Domain Generative Augmentation: Domain Generalization with Latent Diffusion Models
Dec 08, 2023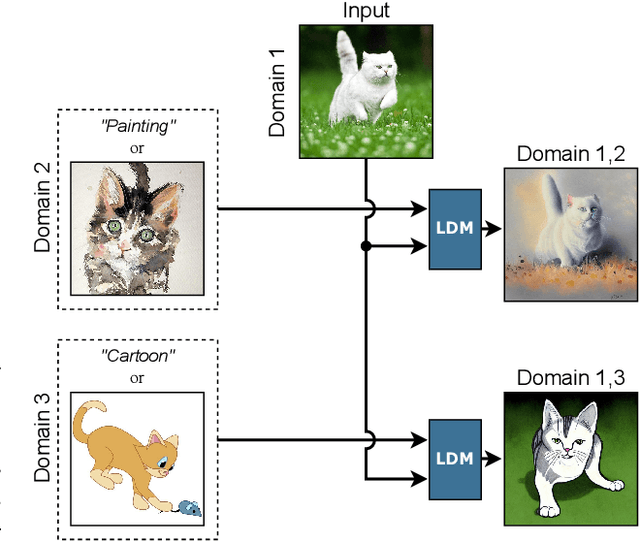



Abstract:Despite the huge effort in developing novel regularizers for Domain Generalization (DG), adding simple data augmentation to the vanilla ERM which is a practical implementation of the Vicinal Risk Minimization principle (VRM) \citep{chapelle2000vicinal} outperforms or stays competitive with many of the proposed regularizers. The VRM reduces the estimation error in ERM by replacing the point-wise kernel estimates with a more precise estimation of true data distribution that reduces the gap between data points \textbf{within each domain}. However, in the DG setting, the estimation error of true data distribution by ERM is mainly caused by the distribution shift \textbf{between domains} which cannot be fully addressed by simple data augmentation techniques within each domain. Inspired by this limitation of VRM, we propose a novel data augmentation named Cross Domain Generative Augmentation (CDGA) that replaces the pointwise kernel estimates in ERM with new density estimates in the \textbf{vicinity of domain pairs} so that the gap between domains is further reduced. To this end, CDGA, which is built upon latent diffusion models (LDM), generates synthetic images to fill the gap between all domains and as a result, reduces the non-iidness. We show that CDGA outperforms SOTA DG methods under the Domainbed benchmark. To explain the effectiveness of CDGA, we generate more than 5 Million synthetic images and perform extensive ablation studies including data scaling laws, distribution visualization, domain shift quantification, adversarial robustness, and loss landscape analysis.
When is a Foundation Model a Foundation Model
Sep 14, 2023

Abstract:Recently, several studies have reported on the fine-tuning of foundation models for image-text modeling in the field of medicine, utilizing images from online data sources such as Twitter and PubMed. Foundation models are large, deep artificial neural networks capable of learning the context of a specific domain through training on exceptionally extensive datasets. Through validation, we have observed that the representations generated by such models exhibit inferior performance in retrieval tasks within digital pathology when compared to those generated by significantly smaller, conventional deep networks.
Understanding Hessian Alignment for Domain Generalization
Aug 22, 2023Abstract:Out-of-distribution (OOD) generalization is a critical ability for deep learning models in many real-world scenarios including healthcare and autonomous vehicles. Recently, different techniques have been proposed to improve OOD generalization. Among these methods, gradient-based regularizers have shown promising performance compared with other competitors. Despite this success, our understanding of the role of Hessian and gradient alignment in domain generalization is still limited. To address this shortcoming, we analyze the role of the classifier's head Hessian matrix and gradient in domain generalization using recent OOD theory of transferability. Theoretically, we show that spectral norm between the classifier's head Hessian matrices across domains is an upper bound of the transfer measure, a notion of distance between target and source domains. Furthermore, we analyze all the attributes that get aligned when we encourage similarity between Hessians and gradients. Our analysis explains the success of many regularizers like CORAL, IRM, V-REx, Fish, IGA, and Fishr as they regularize part of the classifier's head Hessian and/or gradient. Finally, we propose two simple yet effective methods to match the classifier's head Hessians and gradients in an efficient way, based on the Hessian Gradient Product (HGP) and Hutchinson's method (Hutchinson), and without directly calculating Hessians. We validate the OOD generalization ability of proposed methods in different scenarios, including transferability, severe correlation shift, label shift and diversity shift. Our results show that Hessian alignment methods achieve promising performance on various OOD benchmarks. The code is available at \url{https://github.com/huawei-noah/Federated-Learning/tree/main/HessianAlignment}.
Mathematical Challenges in Deep Learning
Mar 24, 2023



Abstract:Deep models are dominating the artificial intelligence (AI) industry since the ImageNet challenge in 2012. The size of deep models is increasing ever since, which brings new challenges to this field with applications in cell phones, personal computers, autonomous cars, and wireless base stations. Here we list a set of problems, ranging from training, inference, generalization bound, and optimization with some formalism to communicate these challenges with mathematicians, statisticians, and theoretical computer scientists. This is a subjective view of the research questions in deep learning that benefits the tech industry in long run.
Learning Binary and Sparse Permutation-Invariant Representations for Fast and Memory Efficient Whole Slide Image Search
Aug 29, 2022


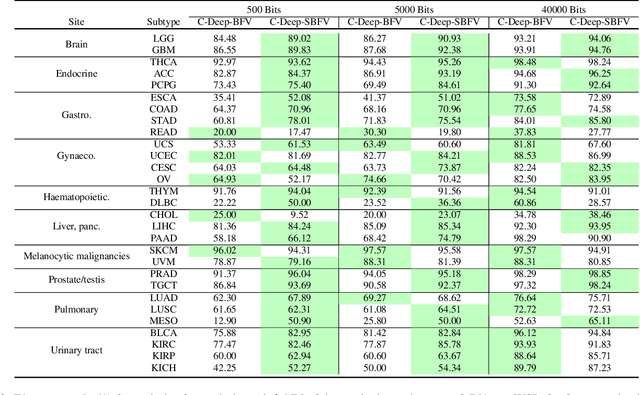
Abstract:Learning suitable Whole slide images (WSIs) representations for efficient retrieval systems is a non-trivial task. The WSI embeddings obtained from current methods are in Euclidean space not ideal for efficient WSI retrieval. Furthermore, most of the current methods require high GPU memory due to the simultaneous processing of multiple sets of patches. To address these challenges, we propose a novel framework for learning binary and sparse WSI representations utilizing a deep generative modelling and the Fisher Vector. We introduce new loss functions for learning sparse and binary permutation-invariant WSI representations that employ instance-based training achieving better memory efficiency. The learned WSI representations are validated on The Cancer Genomic Atlas (TCGA) and Liver-Kidney-Stomach (LKS) datasets. The proposed method outperforms Yottixel (a recent search engine for histopathology images) both in terms of retrieval accuracy and speed. Further, we achieve competitive performance against SOTA on the public benchmark LKS dataset for WSI classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge