H. R. Tizhoosh
Kimia Lab, University of Waterloo, Waterloo, ON, Canada, Vector Institute, MaRS Centre, Toronto, Canada
Aggregation Schemes for Single-Vector WSI Representation Learning in Digital Pathology
Jan 29, 2025



Abstract:A crucial step to efficiently integrate Whole Slide Images (WSIs) in computational pathology is assigning a single high-quality feature vector, i.e., one embedding, to each WSI. With the existence of many pre-trained deep neural networks and the emergence of foundation models, extracting embeddings for sub-images (i.e., tiles or patches) is straightforward. However, for WSIs, given their high resolution and gigapixel nature, inputting them into existing GPUs as a single image is not feasible. As a result, WSIs are usually split into many patches. Feeding each patch to a pre-trained model, each WSI can then be represented by a set of patches, hence, a set of embeddings. Hence, in such a setup, WSI representation learning reduces to set representation learning where for each WSI we have access to a set of patch embeddings. To obtain a single embedding from a set of patch embeddings for each WSI, multiple set-based learning schemes have been proposed in the literature. In this paper, we evaluate the WSI search performance of multiple recently developed aggregation techniques (mainly set representation learning techniques) including simple average or max pooling operations, Deep Sets, Memory networks, Focal attention, Gaussian Mixture Model (GMM) Fisher Vector, and deep sparse and binary Fisher Vector on four different primary sites including bladder, breast, kidney, and Colon from TCGA. Further, we benchmark the search performance of these methods against the median of minimum distances of patch embeddings, a non-aggregating approach used for WSI retrieval.
Zero-Shot Whole Slide Image Retrieval in Histopathology Using Embeddings of Foundation Models
Sep 06, 2024Abstract:We have tested recently published foundation models for histopathology for image retrieval. We report macro average of F1 score for top-1 retrieval, majority of top-3 retrievals, and majority of top-5 retrievals. We perform zero-shot retrievals, i.e., we do not alter embeddings and we do not train any classifier. As test data, we used diagnostic slides of TCGA, The Cancer Genome Atlas, consisting of 23 organs and 117 cancer subtypes. As a search platform we used Yottixel that enabled us to perform WSI search using patches. Achieved F1 scores show low performance, e.g., for top-5 retrievals, 27% +/- 13% (Yottixel-DenseNet), 42% +/- 14% (Yottixel-UNI), 40%+/-13% (Yottixel-Virchow), and 41%+/-13% (Yottixel-GigaPath). The results for GigaPath WSI will be delayed due to the significant computational resources required for processing
A Short Survey on Set-Based Aggregation Techniques for Single-Vector WSI Representation in Digital Pathology
Sep 06, 2024Abstract:Digital pathology is revolutionizing the field of pathology by enabling the digitization, storage, and analysis of tissue samples as whole slide images (WSIs). WSIs are gigapixel files that capture the intricate details of tissue samples, providing a rich source of information for diagnostic and research purposes. However, due to their enormous size, representing these images as one compact vector is essential for many computational pathology tasks, such as search and retrieval, to ensure efficiency and scalability. Most current methods are "patch-oriented," meaning they divide WSIs into smaller patches for processing, which prevents a holistic analysis of the entire slide. Additionally, the necessity for compact representation is driven by the expensive high-performance storage required for WSIs. Not all hospitals have access to such extensive storage solutions, leading to potential disparities in healthcare quality and accessibility. This paper provides an overview of existing set-based approaches to single-vector WSI representation, highlighting the innovations that allow for more efficient and effective use of these complex images in digital pathology, thus addressing both computational challenges and storage limitations.
On Validation of Search & Retrieval of Tissue Images in Digital Pathology
Aug 02, 2024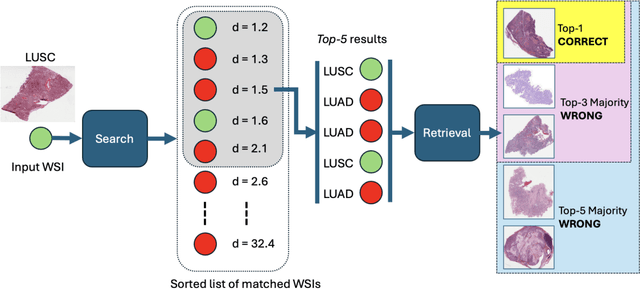


Abstract:Medical images play a crucial role in modern healthcare by providing vital information for diagnosis, treatment planning, and disease monitoring. Fields such as radiology and pathology rely heavily on accurate image interpretation, with radiologists examining X-rays, CT scans, and MRIs to diagnose conditions from fractures to cancer, while pathologists use microscopy and digital images to detect cellular abnormalities for diagnosing cancers and infections. The technological advancements have exponentially increased the volume and complexity of medical images, necessitating efficient tools for management and retrieval. Content-Based Image Retrieval (CBIR) systems address this need by searching and retrieving images based on visual content, enhancing diagnostic accuracy by allowing clinicians to find similar cases and compare pathological patterns. Comprehensive validation of image search engines in medical applications involves evaluating performance metrics like accuracy, indexing, and search times, and storage overhead, ensuring reliable and efficient retrieval of accurate results, as demonstrated by recent validations in histopathology.
SPLICE -- Streamlining Digital Pathology Image Processing
Apr 26, 2024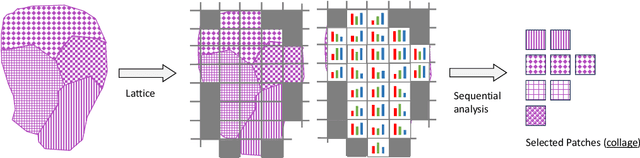
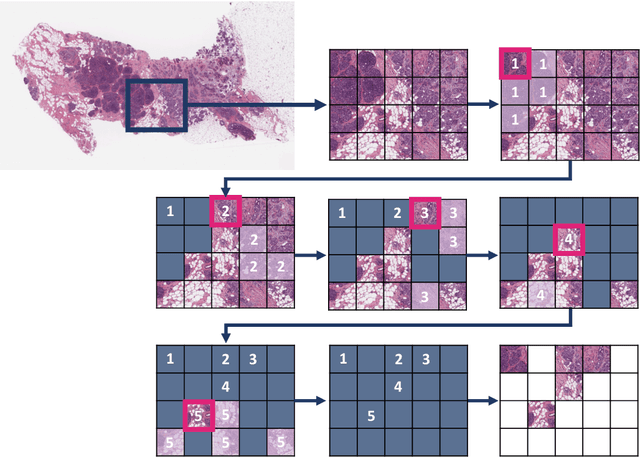
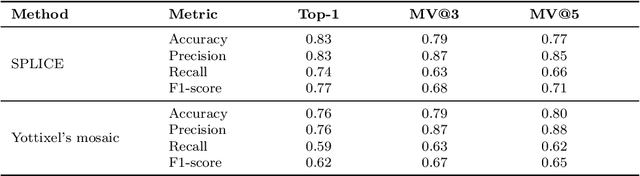
Abstract:Digital pathology and the integration of artificial intelligence (AI) models have revolutionized histopathology, opening new opportunities. With the increasing availability of Whole Slide Images (WSIs), there's a growing demand for efficient retrieval, processing, and analysis of relevant images from vast biomedical archives. However, processing WSIs presents challenges due to their large size and content complexity. Full computer digestion of WSIs is impractical, and processing all patches individually is prohibitively expensive. In this paper, we propose an unsupervised patching algorithm, Sequential Patching Lattice for Image Classification and Enquiry (SPLICE). This novel approach condenses a histopathology WSI into a compact set of representative patches, forming a "collage" of WSI while minimizing redundancy. SPLICE prioritizes patch quality and uniqueness by sequentially analyzing a WSI and selecting non-redundant representative features. We evaluated SPLICE for search and match applications, demonstrating improved accuracy, reduced computation time, and storage requirements compared to existing state-of-the-art methods. As an unsupervised method, SPLICE effectively reduces storage requirements for representing tissue images by 50%. This reduction enables numerous algorithms in computational pathology to operate much more efficiently, paving the way for accelerated adoption of digital pathology.
Foundation Models and Information Retrieval in Digital Pathology
Mar 13, 2024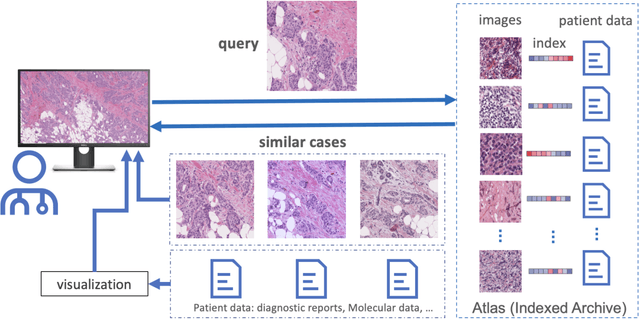
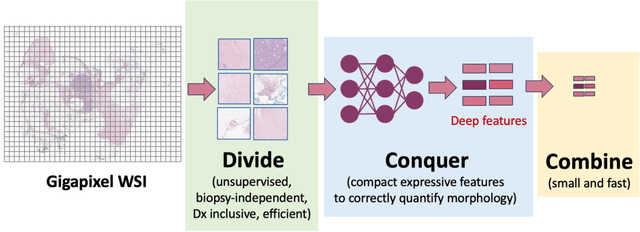
Abstract:The paper reviews the state-of-the-art of foundation models, LLMs, generative AI, information retrieval and CBIR in digital pathology
Training Artificial Neural Networks by Coordinate Search Algorithm
Feb 20, 2024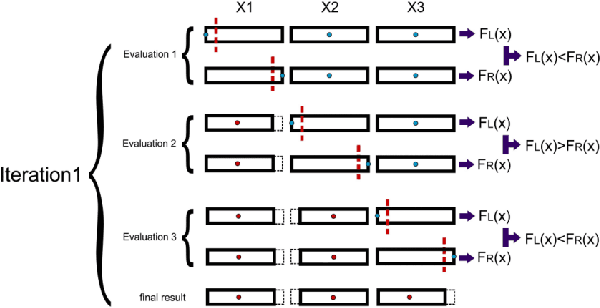
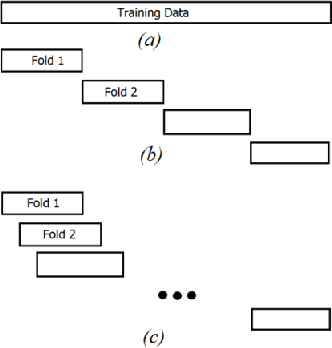
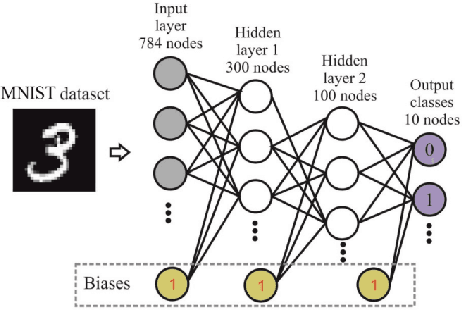
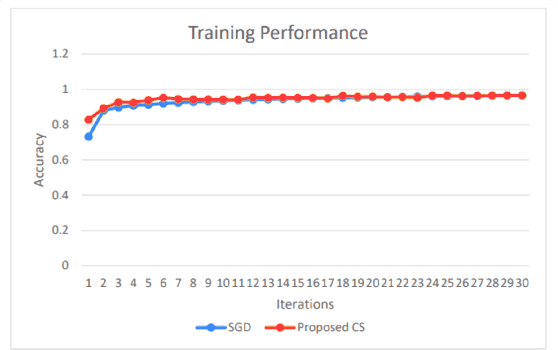
Abstract:Training Artificial Neural Networks poses a challenging and critical problem in machine learning. Despite the effectiveness of gradient-based learning methods, such as Stochastic Gradient Descent (SGD), in training neural networks, they do have several limitations. For instance, they require differentiable activation functions, and cannot optimize a model based on several independent non-differentiable loss functions simultaneously; for example, the F1-score, which is used during testing, can be used during training when a gradient-free optimization algorithm is utilized. Furthermore, the training in any DNN can be possible with a small size of the training dataset. To address these concerns, we propose an efficient version of the gradient-free Coordinate Search (CS) algorithm, an instance of General Pattern Search methods, for training neural networks. The proposed algorithm can be used with non-differentiable activation functions and tailored to multi-objective/multi-loss problems. Finding the optimal values for weights of ANNs is a large-scale optimization problem. Therefore instead of finding the optimal value for each variable, which is the common technique in classical CS, we accelerate optimization and convergence by bundling the weights. In fact, this strategy is a form of dimension reduction for optimization problems. Based on the experimental results, the proposed method, in some cases, outperforms the gradient-based approach, particularly, in situations with insufficient labeled training data. The performance plots demonstrate a high convergence rate, highlighting the capability of our suggested method to find a reasonable solution with fewer function calls. As of now, the only practical and efficient way of training ANNs with hundreds of thousands of weights is gradient-based algorithms such as SGD or Adam. In this paper we introduce an alternative method for training ANN.
* 7 pages, 9 figures
On Image Search in Histopathology
Jan 14, 2024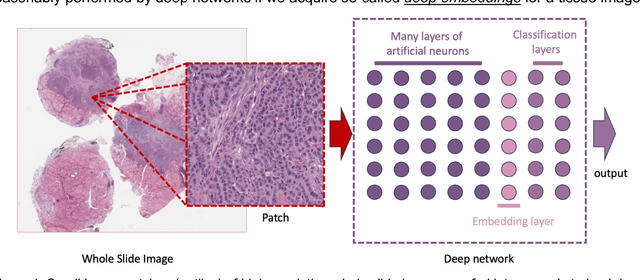
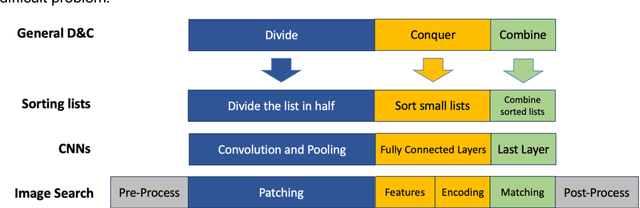
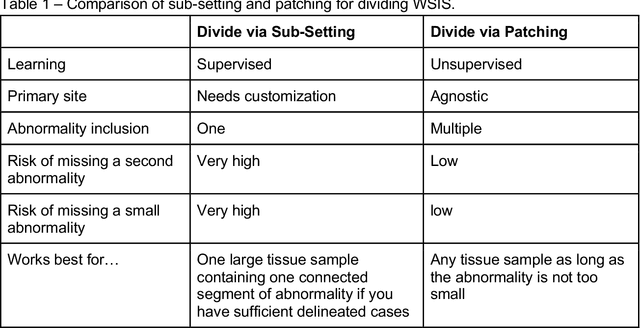
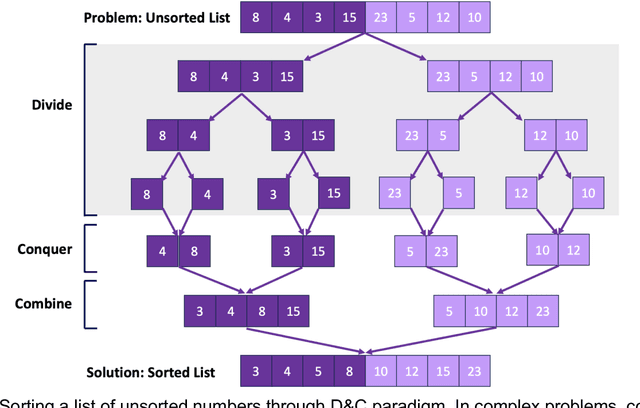
Abstract:Pathology images of histopathology can be acquired from camera-mounted microscopes or whole slide scanners. Utilizing similarity calculations to match patients based on these images holds significant potential in research and clinical contexts. Recent advancements in search technologies allow for nuanced quantification of cellular structures across diverse tissue types, facilitating comparisons and enabling inferences about diagnosis, prognosis, and predictions for new patients when compared against a curated database of diagnosed and treated cases. In this paper, we comprehensively review the latest developments in image search technologies for histopathology, offering a concise overview tailored for computational pathology researchers seeking effective, fast and efficient image search methods in their work.
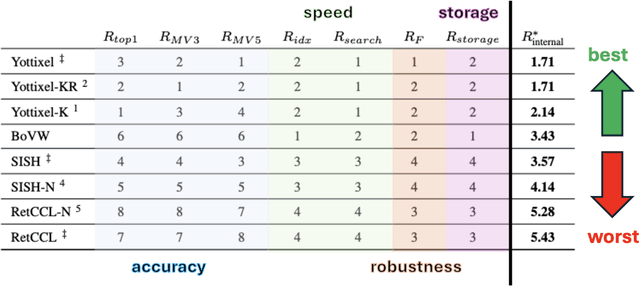
Analysis and Validation of Image Search Engines in Histopathology
Jan 06, 2024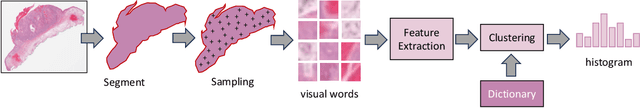
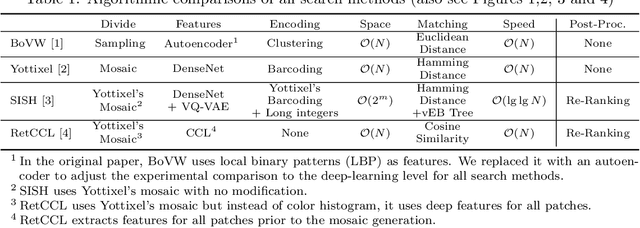
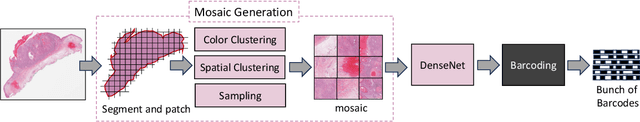
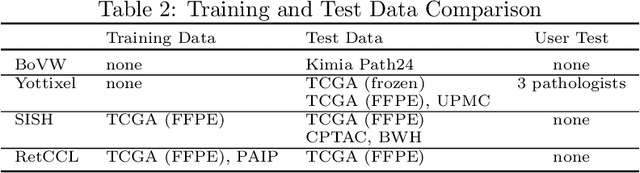
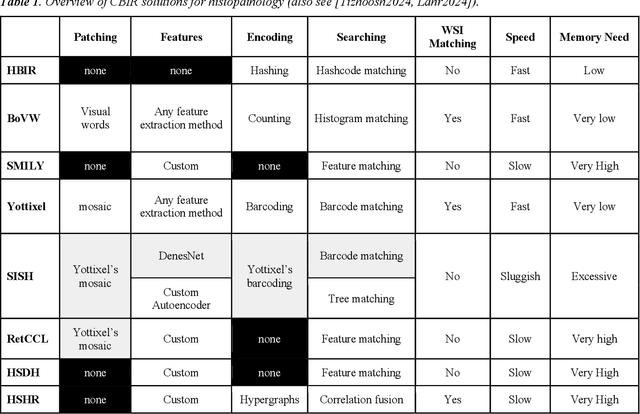
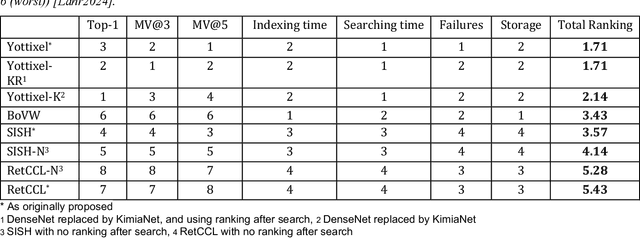
Abstract:Searching for similar images in archives of histology and histopathology images is a crucial task that may aid in patient matching for various purposes, ranging from triaging and diagnosis to prognosis and prediction. Whole slide images (WSIs) are highly detailed digital representations of tissue specimens mounted on glass slides. Matching WSI to WSI can serve as the critical method for patient matching. In this paper, we report extensive analysis and validation of four search methods bag of visual words (BoVW), Yottixel, SISH, RetCCL, and some of their potential variants. We analyze their algorithms and structures and assess their performance. For this evaluation, we utilized four internal datasets ($1269$ patients) and three public datasets ($1207$ patients), totaling more than $200,000$ patches from $38$ different classes/subtypes across five primary sites. Certain search engines, for example, BoVW, exhibit notable efficiency and speed but suffer from low accuracy. Conversely, search engines like Yottixel demonstrate efficiency and speed, providing moderately accurate results. Recent proposals, including SISH, display inefficiency and yield inconsistent outcomes, while alternatives like RetCCL prove inadequate in both accuracy and efficiency. Further research is imperative to address the dual aspects of accuracy and minimal storage requirements in histopathological image search.
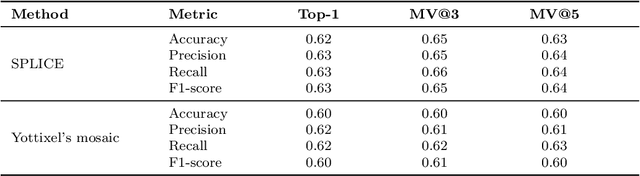
Selection of Distinct Morphologies to Divide & Conquer Gigapixel Pathology Images
Nov 16, 2023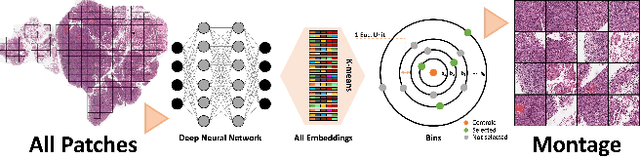
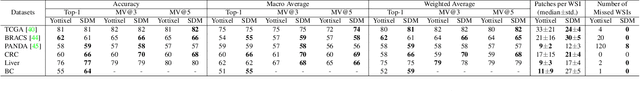
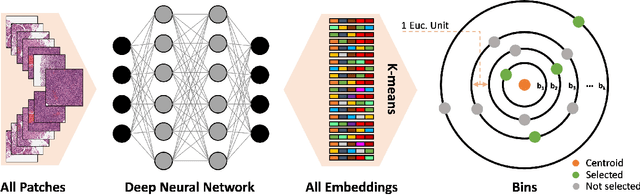
Abstract:Whole slide images (WSIs) are massive digital pathology files illustrating intricate tissue structures. Selecting a small, representative subset of patches from each WSI is essential yet challenging. Therefore, following the "Divide & Conquer" approach becomes essential to facilitate WSI analysis including the classification and the WSI matching in computational pathology. To this end, we propose a novel method termed "Selection of Distinct Morphologies" (SDM) to choose a subset of WSI patches. The aim is to encompass all inherent morphological variations within a given WSI while simultaneously minimizing the number of selected patches to represent these variations, ensuring a compact yet comprehensive set of patches. This systematically curated patch set forms what we term a "montage". We assess the representativeness of the SDM montage across various public and private histopathology datasets. This is conducted by using the leave-one-out WSI search and matching evaluation method, comparing it with the state-of-the-art Yottixel's mosaic. SDM demonstrates remarkable efficacy across all datasets during its evaluation. Furthermore, SDM eliminates the necessity for empirical parameterization, a crucial aspect of Yottixel's mosaic, by inherently optimizing the selection process to capture the distinct morphological features within the WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge