Jibran Khan
SPLICE -- Streamlining Digital Pathology Image Processing
Apr 26, 2024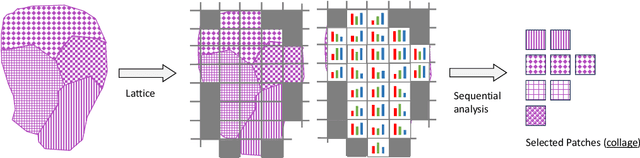
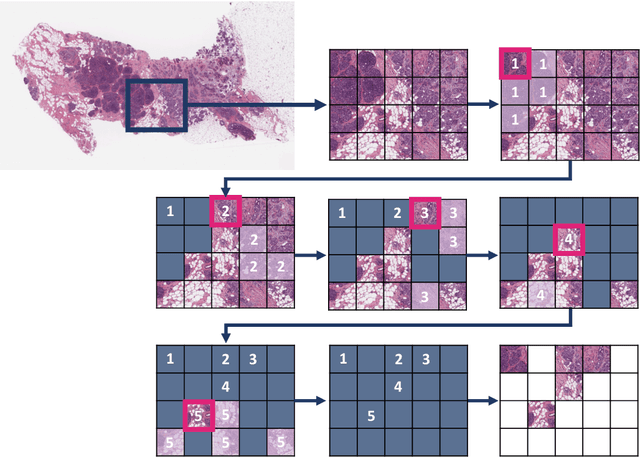
Abstract:Digital pathology and the integration of artificial intelligence (AI) models have revolutionized histopathology, opening new opportunities. With the increasing availability of Whole Slide Images (WSIs), there's a growing demand for efficient retrieval, processing, and analysis of relevant images from vast biomedical archives. However, processing WSIs presents challenges due to their large size and content complexity. Full computer digestion of WSIs is impractical, and processing all patches individually is prohibitively expensive. In this paper, we propose an unsupervised patching algorithm, Sequential Patching Lattice for Image Classification and Enquiry (SPLICE). This novel approach condenses a histopathology WSI into a compact set of representative patches, forming a "collage" of WSI while minimizing redundancy. SPLICE prioritizes patch quality and uniqueness by sequentially analyzing a WSI and selecting non-redundant representative features. We evaluated SPLICE for search and match applications, demonstrating improved accuracy, reduced computation time, and storage requirements compared to existing state-of-the-art methods. As an unsupervised method, SPLICE effectively reduces storage requirements for representing tissue images by 50%. This reduction enables numerous algorithms in computational pathology to operate much more efficiently, paving the way for accelerated adoption of digital pathology.
Analysis and Validation of Image Search Engines in Histopathology
Jan 06, 2024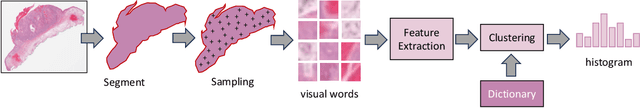
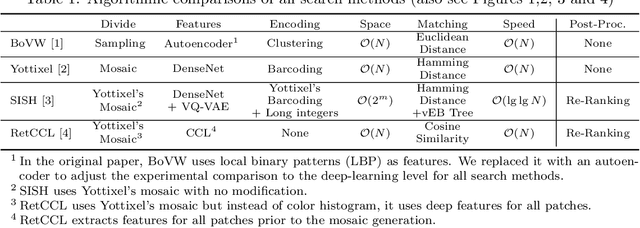
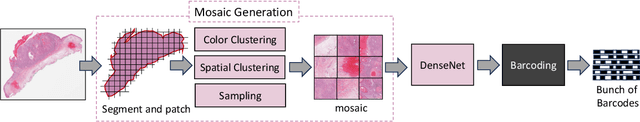
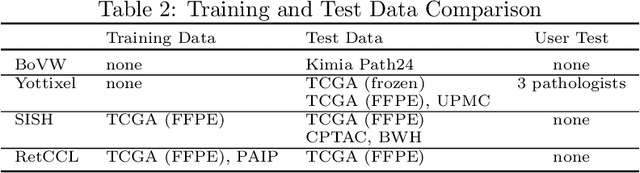
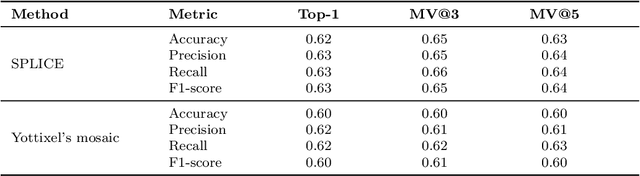
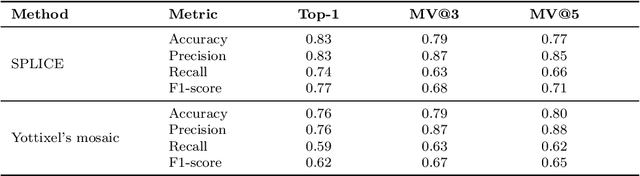
Abstract:Searching for similar images in archives of histology and histopathology images is a crucial task that may aid in patient matching for various purposes, ranging from triaging and diagnosis to prognosis and prediction. Whole slide images (WSIs) are highly detailed digital representations of tissue specimens mounted on glass slides. Matching WSI to WSI can serve as the critical method for patient matching. In this paper, we report extensive analysis and validation of four search methods bag of visual words (BoVW), Yottixel, SISH, RetCCL, and some of their potential variants. We analyze their algorithms and structures and assess their performance. For this evaluation, we utilized four internal datasets ($1269$ patients) and three public datasets ($1207$ patients), totaling more than $200,000$ patches from $38$ different classes/subtypes across five primary sites. Certain search engines, for example, BoVW, exhibit notable efficiency and speed but suffer from low accuracy. Conversely, search engines like Yottixel demonstrate efficiency and speed, providing moderately accurate results. Recent proposals, including SISH, display inefficiency and yield inconsistent outcomes, while alternatives like RetCCL prove inadequate in both accuracy and efficiency. Further research is imperative to address the dual aspects of accuracy and minimal storage requirements in histopathological image search.
Rotation-Agnostic Image Representation Learning for Digital Pathology
Nov 14, 2023

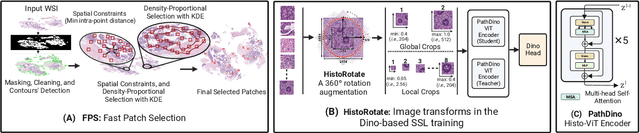
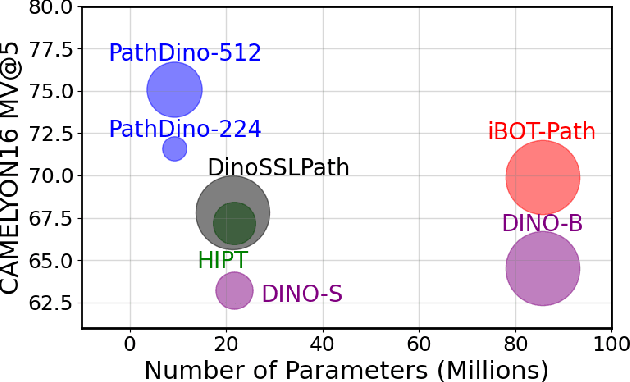
Abstract:This paper addresses complex challenges in histopathological image analysis through three key contributions. Firstly, it introduces a fast patch selection method, FPS, for whole-slide image (WSI) analysis, significantly reducing computational cost while maintaining accuracy. Secondly, it presents PathDino, a lightweight histopathology feature extractor with a minimal configuration of five Transformer blocks and only 9 million parameters, markedly fewer than alternatives. Thirdly, it introduces a rotation-agnostic representation learning paradigm using self-supervised learning, effectively mitigating overfitting. We also show that our compact model outperforms existing state-of-the-art histopathology-specific vision transformers on 12 diverse datasets, including both internal datasets spanning four sites (breast, liver, skin, and colorectal) and seven public datasets (PANDA, CAMELYON16, BRACS, DigestPath, Kather, PanNuke, and WSSS4LUAD). Notably, even with a training dataset of 6 million histopathology patches from The Cancer Genome Atlas (TCGA), our approach demonstrates an average 8.5% improvement in patch-level majority vote performance. These contributions provide a robust framework for enhancing image analysis in digital pathology, rigorously validated through extensive evaluation. Project Page: https://rhazeslab.github.io/PathDino-Page/
When is a Foundation Model a Foundation Model
Sep 14, 2023

Abstract:Recently, several studies have reported on the fine-tuning of foundation models for image-text modeling in the field of medicine, utilizing images from online data sources such as Twitter and PubMed. Foundation models are large, deep artificial neural networks capable of learning the context of a specific domain through training on exceptionally extensive datasets. Through validation, we have observed that the representations generated by such models exhibit inferior performance in retrieval tasks within digital pathology when compared to those generated by significantly smaller, conventional deep networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge