Dennis Murphree
Creating an Atlas of Normal Tissue for Pruning WSI Patching Through Anomaly Detection
Oct 04, 2023Abstract:Patching gigapixel whole slide images (WSIs) is an important task in computational pathology. Some methods have been proposed to select a subset of patches as WSI representation for downstream tasks. While most of the computational pathology tasks are designed to classify or detect the presence of pathological lesions in each WSI, the confounding role and redundant nature of normal histology in tissue samples are generally overlooked in WSI representations. In this paper, we propose and validate the concept of an "atlas of normal tissue" solely using samples of WSIs obtained from normal tissue biopsies. Such atlases can be employed to eliminate normal fragments of tissue samples and hence increase the representativeness collection of patches. We tested our proposed method by establishing a normal atlas using 107 normal skin WSIs and demonstrated how established indexes and search engines like Yottixel can be improved. We used 553 WSIs of cutaneous squamous cell carcinoma (cSCC) to show the advantage. We also validated our method applied to an external dataset of 451 breast WSIs. The number of selected WSI patches was reduced by 30% to 50% after utilizing the proposed normal atlas while maintaining the same indexing and search performance in leave-one-patinet-out validation for both datasets. We show that the proposed normal atlas shows promise for unsupervised selection of the most representative patches of the abnormal/malignant WSI lesions.
When is a Foundation Model a Foundation Model
Sep 14, 2023

Abstract:Recently, several studies have reported on the fine-tuning of foundation models for image-text modeling in the field of medicine, utilizing images from online data sources such as Twitter and PubMed. Foundation models are large, deep artificial neural networks capable of learning the context of a specific domain through training on exceptionally extensive datasets. Through validation, we have observed that the representations generated by such models exhibit inferior performance in retrieval tasks within digital pathology when compared to those generated by significantly smaller, conventional deep networks.
RACR-MIL: Weakly Supervised Skin Cancer Grading using Rank-Aware Contextual Reasoning on Whole Slide Images
Aug 29, 2023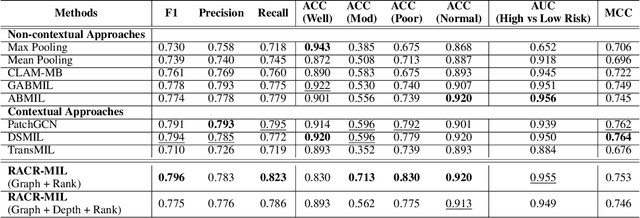
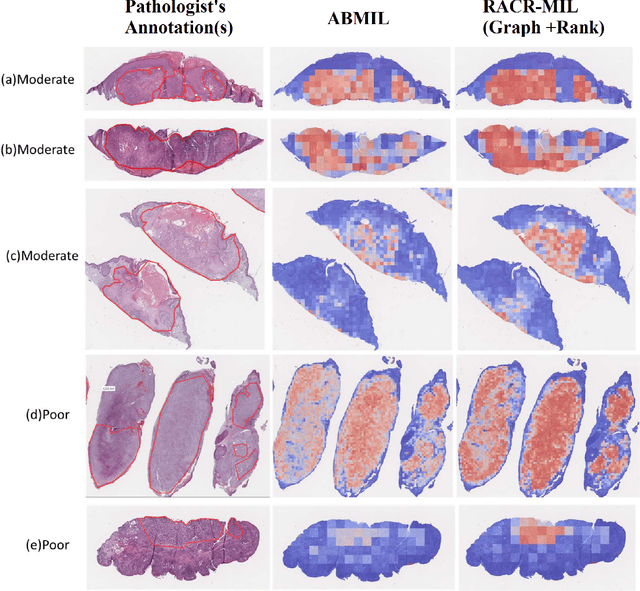
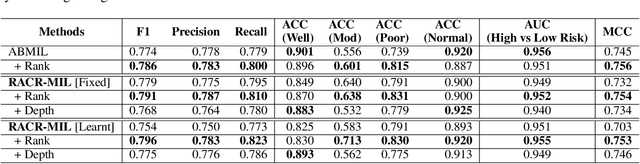
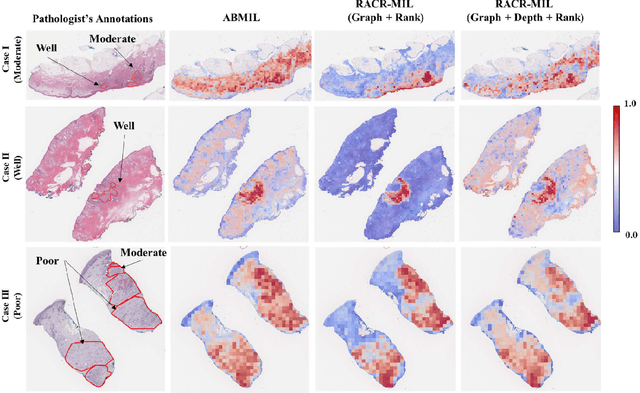
Abstract:Cutaneous squamous cell cancer (cSCC) is the second most common skin cancer in the US. It is diagnosed by manual multi-class tumor grading using a tissue whole slide image (WSI), which is subjective and suffers from inter-pathologist variability. We propose an automated weakly-supervised grading approach for cSCC WSIs that is trained using WSI-level grade and does not require fine-grained tumor annotations. The proposed model, RACR-MIL, transforms each WSI into a bag of tiled patches and leverages attention-based multiple-instance learning to assign a WSI-level grade. We propose three key innovations to address general as well as cSCC-specific challenges in tumor grading. First, we leverage spatial and semantic proximity to define a WSI graph that encodes both local and non-local dependencies between tumor regions and leverage graph attention convolution to derive contextual patch features. Second, we introduce a novel ordinal ranking constraint on the patch attention network to ensure that higher-grade tumor regions are assigned higher attention. Third, we use tumor depth as an auxiliary task to improve grade classification in a multitask learning framework. RACR-MIL achieves 2-9% improvement in grade classification over existing weakly-supervised approaches on a dataset of 718 cSCC tissue images and localizes the tumor better. The model achieves 5-20% higher accuracy in difficult-to-classify high-risk grade classes and is robust to class imbalance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge