Azam Asilian Bidgoli
EvoPS: Evolutionary Patch Selection for Whole Slide Image Analysis in Computational Pathology
Nov 10, 2025Abstract:In computational pathology, the gigapixel scale of Whole-Slide Images (WSIs) necessitates their division into thousands of smaller patches. Analyzing these high-dimensional patch embeddings is computationally expensive and risks diluting key diagnostic signals with many uninformative patches. Existing patch selection methods often rely on random sampling or simple clustering heuristics and typically fail to explicitly manage the crucial trade-off between the number of selected patches and the accuracy of the resulting slide representation. To address this gap, we propose EvoPS (Evolutionary Patch Selection), a novel framework that formulates patch selection as a multi-objective optimization problem and leverages an evolutionary search to simultaneously minimize the number of selected patch embeddings and maximize the performance of a downstream similarity search task, generating a Pareto front of optimal trade-off solutions. We validated our framework across four major cancer cohorts from The Cancer Genome Atlas (TCGA) using five pretrained deep learning models to generate patch embeddings, including both supervised CNNs and large self-supervised foundation models. The results demonstrate that EvoPS can reduce the required number of training patch embeddings by over 90% while consistently maintaining or even improving the final classification F1-score compared to a baseline that uses all available patches' embeddings selected through a standard extraction pipeline. The EvoPS framework provides a robust and principled method for creating efficient, accurate, and interpretable WSI representations, empowering users to select an optimal balance between computational cost and diagnostic performance.
Evolutionary Feature-wise Thresholding for Binary Representation of NLP Embeddings
Jul 22, 2025Abstract:Efficient text embedding is crucial for large-scale natural language processing (NLP) applications, where storage and computational efficiency are key concerns. In this paper, we explore how using binary representations (barcodes) instead of real-valued features can be used for NLP embeddings derived from machine learning models such as BERT. Thresholding is a common method for converting continuous embeddings into binary representations, often using a fixed threshold across all features. We propose a Coordinate Search-based optimization framework that instead identifies the optimal threshold for each feature, demonstrating that feature-specific thresholds lead to improved performance in binary encoding. This ensures that the binary representations are both accurate and efficient, enhancing performance across various features. Our optimal barcode representations have shown promising results in various NLP applications, demonstrating their potential to transform text representation. We conducted extensive experiments and statistical tests on different NLP tasks and datasets to evaluate our approach and compare it to other thresholding methods. Binary embeddings generated using using optimal thresholds found by our method outperform traditional binarization methods in accuracy. This technique for generating binary representations is versatile and can be applied to any features, not just limited to NLP embeddings, making it useful for a wide range of domains in machine learning applications.
A Novel Pareto-optimal Ranking Method for Comparing Multi-objective Optimization Algorithms
Nov 27, 2024



Abstract:As the interest in multi- and many-objective optimization algorithms grows, the performance comparison of these algorithms becomes increasingly important. A large number of performance indicators for multi-objective optimization algorithms have been introduced, each of which evaluates these algorithms based on a certain aspect. Therefore, assessing the quality of multi-objective results using multiple indicators is essential to guarantee that the evaluation considers all quality perspectives. This paper proposes a novel multi-metric comparison method to rank the performance of multi-/ many-objective optimization algorithms based on a set of performance indicators. We utilize the Pareto optimality concept (i.e., non-dominated sorting algorithm) to create the rank levels of algorithms by simultaneously considering multiple performance indicators as criteria/objectives. As a result, four different techniques are proposed to rank algorithms based on their contribution at each Pareto level. This method allows researchers to utilize a set of existing/newly developed performance metrics to adequately assess/rank multi-/many-objective algorithms. The proposed methods are scalable and can accommodate in its comprehensive scheme any newly introduced metric. The method was applied to rank 10 competing algorithms in the 2018 CEC competition solving 15 many-objective test problems. The Pareto-optimal ranking was conducted based on 10 well-known multi-objective performance indicators and the results were compared to the final ranks reported by the competition, which were based on the inverted generational distance (IGD) and hypervolume indicator (HV) measures. The techniques suggested in this paper have broad applications in science and engineering, particularly in areas where multiple metrics are used for comparisons. Examples include machine learning and data mining.
Enhancing Diversity in Multi-objective Feature Selection
Jul 25, 2024Abstract:Feature selection plays a pivotal role in the data preprocessing and model-building pipeline, significantly enhancing model performance, interpretability, and resource efficiency across diverse domains. In population-based optimization methods, the generation of diverse individuals holds utmost importance for adequately exploring the problem landscape, particularly in highly multi-modal multi-objective optimization problems. Our study reveals that, in line with findings from several prior research papers, commonly employed crossover and mutation operations lack the capability to generate high-quality diverse individuals and tend to become confined to limited areas around various local optima. This paper introduces an augmentation to the diversity of the population in the well-established multi-objective scheme of the genetic algorithm, NSGA-II. This enhancement is achieved through two key components: the genuine initialization method and the substitution of the worst individuals with new randomly generated individuals as a re-initialization approach in each generation. The proposed multi-objective feature selection method undergoes testing on twelve real-world classification problems, with the number of features ranging from 2,400 to nearly 50,000. The results demonstrate that replacing the last front of the population with an equivalent number of new random individuals generated using the genuine initialization method and featuring a limited number of features substantially improves the population's quality and, consequently, enhances the performance of the multi-objective algorithm.
Training Artificial Neural Networks by Coordinate Search Algorithm
Feb 20, 2024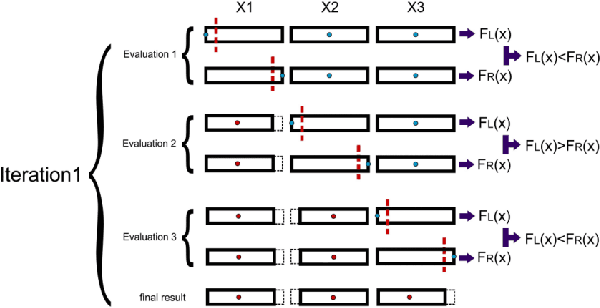
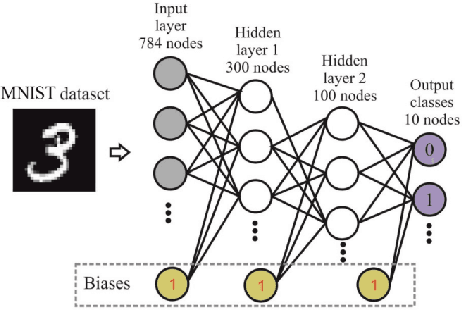
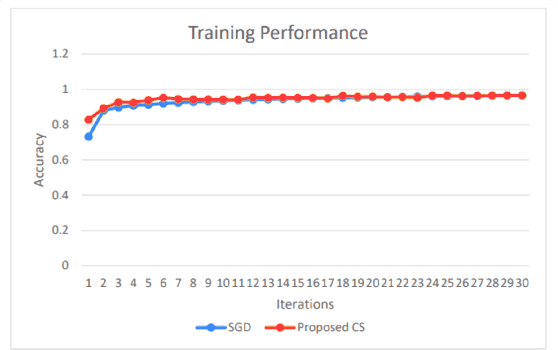
Abstract:Training Artificial Neural Networks poses a challenging and critical problem in machine learning. Despite the effectiveness of gradient-based learning methods, such as Stochastic Gradient Descent (SGD), in training neural networks, they do have several limitations. For instance, they require differentiable activation functions, and cannot optimize a model based on several independent non-differentiable loss functions simultaneously; for example, the F1-score, which is used during testing, can be used during training when a gradient-free optimization algorithm is utilized. Furthermore, the training in any DNN can be possible with a small size of the training dataset. To address these concerns, we propose an efficient version of the gradient-free Coordinate Search (CS) algorithm, an instance of General Pattern Search methods, for training neural networks. The proposed algorithm can be used with non-differentiable activation functions and tailored to multi-objective/multi-loss problems. Finding the optimal values for weights of ANNs is a large-scale optimization problem. Therefore instead of finding the optimal value for each variable, which is the common technique in classical CS, we accelerate optimization and convergence by bundling the weights. In fact, this strategy is a form of dimension reduction for optimization problems. Based on the experimental results, the proposed method, in some cases, outperforms the gradient-based approach, particularly, in situations with insufficient labeled training data. The performance plots demonstrate a high convergence rate, highlighting the capability of our suggested method to find a reasonable solution with fewer function calls. As of now, the only practical and efficient way of training ANNs with hundreds of thousands of weights is gradient-based algorithms such as SGD or Adam. In this paper we introduce an alternative method for training ANN.
* 7 pages, 9 figures
Compact NSGA-II for Multi-objective Feature Selection
Feb 20, 2024



Abstract:Feature selection is an expensive challenging task in machine learning and data mining aimed at removing irrelevant and redundant features. This contributes to an improvement in classification accuracy, as well as the budget and memory requirements for classification, or any other post-processing task conducted after feature selection. In this regard, we define feature selection as a multi-objective binary optimization task with the objectives of maximizing classification accuracy and minimizing the number of selected features. In order to select optimal features, we have proposed a binary Compact NSGA-II (CNSGA-II) algorithm. Compactness represents the population as a probability distribution to enhance evolutionary algorithms not only to be more memory-efficient but also to reduce the number of fitness evaluations. Instead of holding two populations during the optimization process, our proposed method uses several Probability Vectors (PVs) to generate new individuals. Each PV efficiently explores a region of the search space to find non-dominated solutions instead of generating candidate solutions from a small population as is the common approach in most evolutionary algorithms. To the best of our knowledge, this is the first compact multi-objective algorithm proposed for feature selection. The reported results for expensive optimization cases with a limited budget on five datasets show that the CNSGA-II performs more efficiently than the well-known NSGA-II method in terms of the hypervolume (HV) performance metric requiring less memory. The proposed method and experimental results are explained and analyzed in detail.
* 8 pages, 2 figures
Multi-objective Binary Coordinate Search for Feature Selection
Feb 20, 2024



Abstract:A supervised feature selection method selects an appropriate but concise set of features to differentiate classes, which is highly expensive for large-scale datasets. Therefore, feature selection should aim at both minimizing the number of selected features and maximizing the accuracy of classification, or any other task. However, this crucial task is computationally highly demanding on many real-world datasets and requires a very efficient algorithm to reach a set of optimal features with a limited number of fitness evaluations. For this purpose, we have proposed the binary multi-objective coordinate search (MOCS) algorithm to solve large-scale feature selection problems. To the best of our knowledge, the proposed algorithm in this paper is the first multi-objective coordinate search algorithm. In this method, we generate new individuals by flipping a variable of the candidate solutions on the Pareto front. This enables us to investigate the effectiveness of each feature in the corresponding subset. In fact, this strategy can play the role of crossover and mutation operators to generate distinct subsets of features. The reported results indicate the significant superiority of our method over NSGA-II, on five real-world large-scale datasets, particularly when the computing budget is limited. Moreover, this simple hyper-parameter-free algorithm can solve feature selection much faster and more efficiently than NSGA-II.
* 8 pages, 1 figure
Ranking Loss and Sequestering Learning for Reducing Image Search Bias in Histopathology
Apr 15, 2023Abstract:Recently, deep learning has started to play an essential role in healthcare applications, including image search in digital pathology. Despite the recent progress in computer vision, significant issues remain for image searching in histopathology archives. A well-known problem is AI bias and lack of generalization. A more particular shortcoming of deep models is the ignorance toward search functionality. The former affects every model, the latter only search and matching. Due to the lack of ranking-based learning, researchers must train models based on the classification error and then use the resultant embedding for image search purposes. Moreover, deep models appear to be prone to internal bias even if using a large image repository of various hospitals. This paper proposes two novel ideas to improve image search performance. First, we use a ranking loss function to guide feature extraction toward the matching-oriented nature of the search. By forcing the model to learn the ranking of matched outputs, the representation learning is customized toward image search instead of learning a class label. Second, we introduce the concept of sequestering learning to enhance the generalization of feature extraction. By excluding the images of the input hospital from the matched outputs, i.e., sequestering the input domain, the institutional bias is reduced. The proposed ideas are implemented and validated through the largest public dataset of whole slide images. The experiments demonstrate superior results compare to the-state-of-art.
Evolutionary Computation in Action: Hyperdimensional Deep Embedding Spaces of Gigapixel Pathology Images
Mar 02, 2023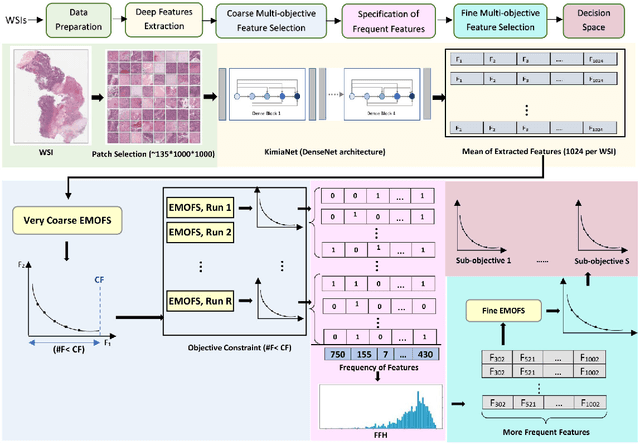



Abstract:One of the main obstacles of adopting digital pathology is the challenge of efficient processing of hyperdimensional digitized biopsy samples, called whole slide images (WSIs). Exploiting deep learning and introducing compact WSI representations are urgently needed to accelerate image analysis and facilitate the visualization and interpretability of pathology results in a postpandemic world. In this paper, we introduce a new evolutionary approach for WSI representation based on large-scale multi-objective optimization (LSMOP) of deep embeddings. We start with patch-based sampling to feed KimiaNet , a histopathology-specialized deep network, and to extract a multitude of feature vectors. Coarse multi-objective feature selection uses the reduced search space strategy guided by the classification accuracy and the number of features. In the second stage, the frequent features histogram (FFH), a novel WSI representation, is constructed by multiple runs of coarse LSMOP. Fine evolutionary feature selection is then applied to find a compact (short-length) feature vector based on the FFH and contributes to a more robust deep-learning approach to digital pathology supported by the stochastic power of evolutionary algorithms. We validate the proposed schemes using The Cancer Genome Atlas (TCGA) images in terms of WSI representation, classification accuracy, and feature quality. Furthermore, a novel decision space for multicriteria decision making in the LSMOP field is introduced. Finally, a patch-level visualization approach is proposed to increase the interpretability of deep features. The proposed evolutionary algorithm finds a very compact feature vector to represent a WSI (almost 14,000 times smaller than the original feature vectors) with 8% higher accuracy compared to the codes provided by the state-of-the-art methods.
Image-Based Benchmarking and Visualization for Large-Scale Global Optimization
Jul 24, 2020



Abstract:In the context of optimization, visualization techniques can be useful for understanding the behaviour of optimization algorithms and can even provide a means to facilitate human interaction with an optimizer. Towards this goal, an image-based visualization framework, without dimension reduction, that visualizes the solutions to large-scale global optimization problems as images is proposed. In the proposed framework, the pixels visualize decision variables while the entire image represents the overall solution quality. This framework affords a number of benefits over existing visualization techniques including enhanced scalability (in terms of the number of decision variables), facilitation of standard image processing techniques, providing nearly infinite benchmark cases, and explicit alignment with human perception. Furthermore, image-based visualization can be used to visualize the optimization process in real-time, thereby allowing the user to ascertain characteristics of the search process as it is progressing. To the best of the authors' knowledge, this is the first realization of a dimension-preserving, scalable visualization framework that embeds the inherent relationship between decision space and objective space. The proposed framework is utilized with 10 different mapping schemes on an image-reconstruction problem that encompass continuous, discrete, binary, combinatorial, constrained, dynamic, and multi-objective optimization. The proposed framework is then demonstrated on arbitrary benchmark problems with known optima. Experimental results elucidate the flexibility and demonstrate how valuable information about the search process can be gathered via the proposed visualization framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge