Abtin Riasatian
Evolutionary Computation in Action: Hyperdimensional Deep Embedding Spaces of Gigapixel Pathology Images
Mar 02, 2023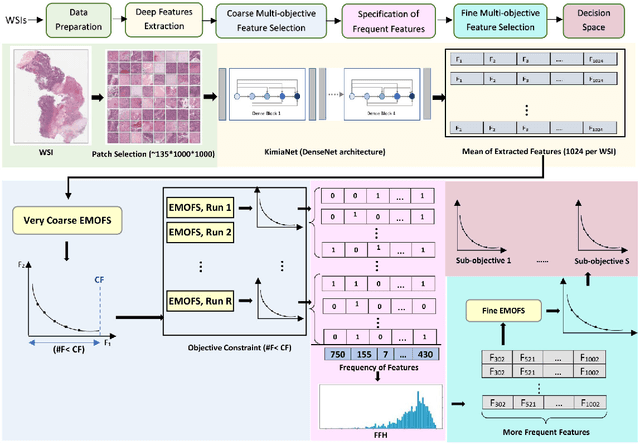



Abstract:One of the main obstacles of adopting digital pathology is the challenge of efficient processing of hyperdimensional digitized biopsy samples, called whole slide images (WSIs). Exploiting deep learning and introducing compact WSI representations are urgently needed to accelerate image analysis and facilitate the visualization and interpretability of pathology results in a postpandemic world. In this paper, we introduce a new evolutionary approach for WSI representation based on large-scale multi-objective optimization (LSMOP) of deep embeddings. We start with patch-based sampling to feed KimiaNet , a histopathology-specialized deep network, and to extract a multitude of feature vectors. Coarse multi-objective feature selection uses the reduced search space strategy guided by the classification accuracy and the number of features. In the second stage, the frequent features histogram (FFH), a novel WSI representation, is constructed by multiple runs of coarse LSMOP. Fine evolutionary feature selection is then applied to find a compact (short-length) feature vector based on the FFH and contributes to a more robust deep-learning approach to digital pathology supported by the stochastic power of evolutionary algorithms. We validate the proposed schemes using The Cancer Genome Atlas (TCGA) images in terms of WSI representation, classification accuracy, and feature quality. Furthermore, a novel decision space for multicriteria decision making in the LSMOP field is introduced. Finally, a patch-level visualization approach is proposed to increase the interpretability of deep features. The proposed evolutionary algorithm finds a very compact feature vector to represent a WSI (almost 14,000 times smaller than the original feature vectors) with 8% higher accuracy compared to the codes provided by the state-of-the-art methods.
Gram Barcodes for Histopathology Tissue Texture Retrieval
Nov 28, 2021
Abstract:Recent advances in digital pathology have led to the need for Histopathology Image Retrieval (HIR) systems that search through databases of biopsy images to find similar cases to a given query image. These HIR systems allow pathologists to effortlessly and efficiently access thousands of previously diagnosed cases in order to exploit the knowledge in the corresponding pathology reports. Since HIR systems may have to deal with millions of gigapixel images, the extraction of compact and expressive image features must be available to allow for efficient and accurate retrieval. In this paper, we propose the application of Gram barcodes as image features for HIR systems. Unlike most feature generation schemes, Gram barcodes are based on high-order statistics that describe tissue texture by summarizing the correlations between different feature maps in layers of convolutional neural networks. We run HIR experiments on three public datasets using a pre-trained VGG19 network for Gram barcode generation and showcase highly competitive results.
Searching for Pneumothorax in X-Ray Images Using Autoencoded Deep Features
Feb 11, 2021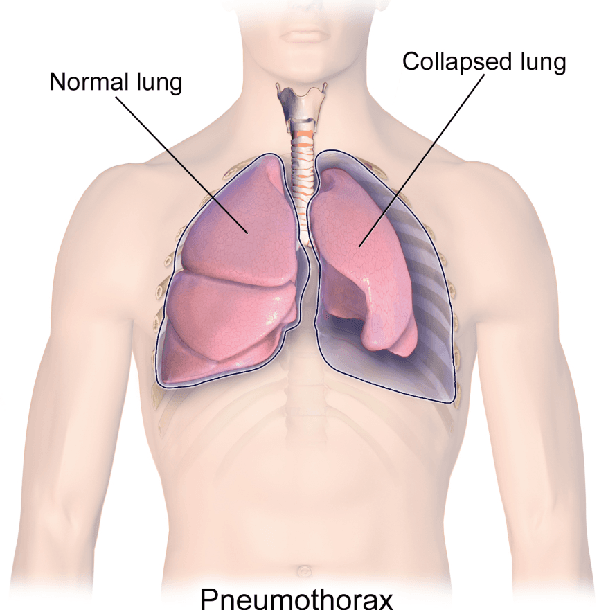

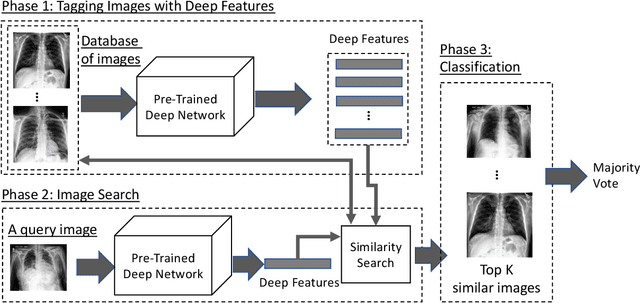
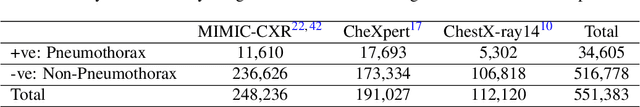
Abstract:Fast diagnosis and treatment of pneumothorax, a collapsed or dropped lung, is crucial to avoid fatalities. Pneumothorax is typically detected on a chest X-ray image through visual inspection by experienced radiologists. However, the detection rate is quite low. Therefore, there is a strong need for automated detection systems to assist radiologists. Despite the high accuracy levels generally reported for deep learning classifiers in many applications, they may not be useful in clinical practice due to the lack of large number of high-quality labelled images as well as a lack of interpretation possibility. Alternatively, searching in the archive of past cases to find matching images may serve as a 'virtual second opinion' through accessing the metadata of matched evidently diagnosed cases. To use image search as a triaging/diagnosis tool, all chest X-ray images must first be tagged with identifiers, i.e., deep features. Then, given a query chest X-ray image, the majority vote among the top k retrieved images can provide a more explainable output. While image search can be clinically more viable, its detection performance needs to be investigated at a scale closer to real-world practice. We combined 3 public datasets to assemble a repository with more than 550,000 chest X-ray images. We developed the Autoencoding Thorax Net (short AutoThorax-Net) for image search in chest radiographs compressing three inputs: the left chest side, the flipped right side, and the entire chest image. Experimental results show that image search based on AutoThorax-Net features can achieve high identification rates providing a path towards real-world deployment. We achieved 92% AUC accuracy for a semi-automated search in 194,608 images (pneumothorax and normal) and 82% AUC accuracy for fully automated search in 551,383 images (normal, pneumothorax and many other chest diseases).
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021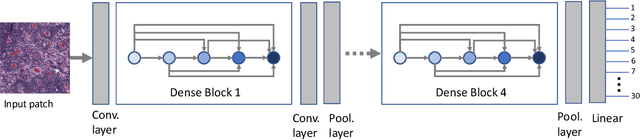
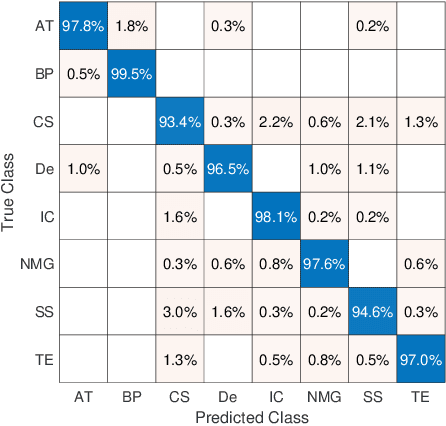
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
A Comparative Study of U-Net Topologies for Background Removal in Histopathology Images
Jun 08, 2020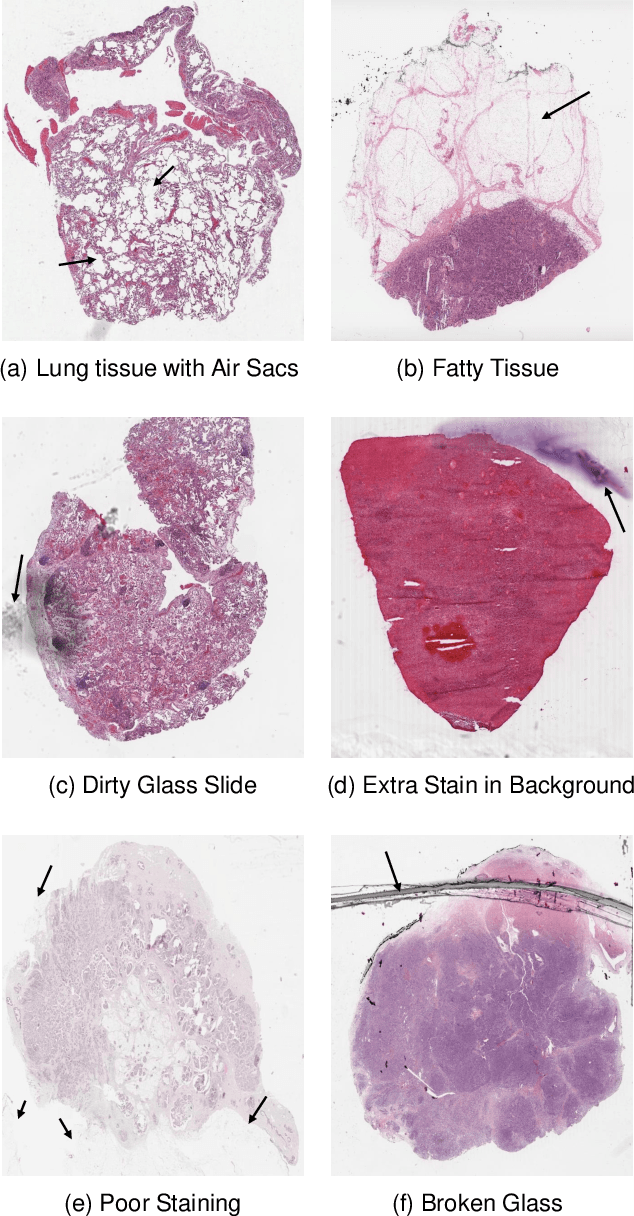
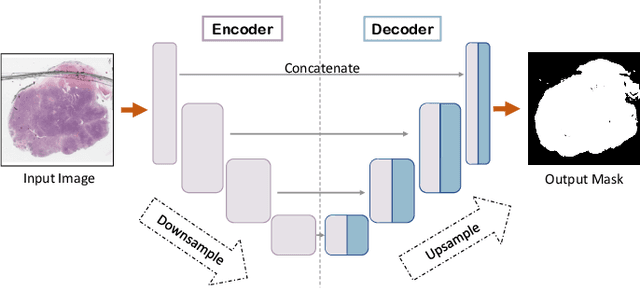
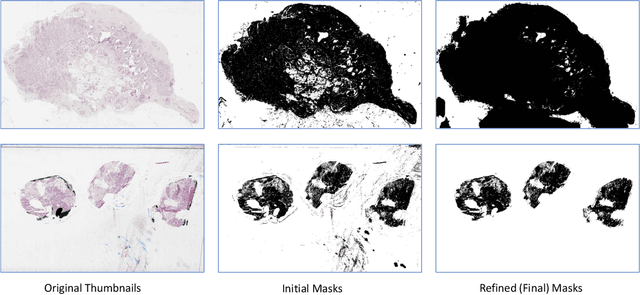
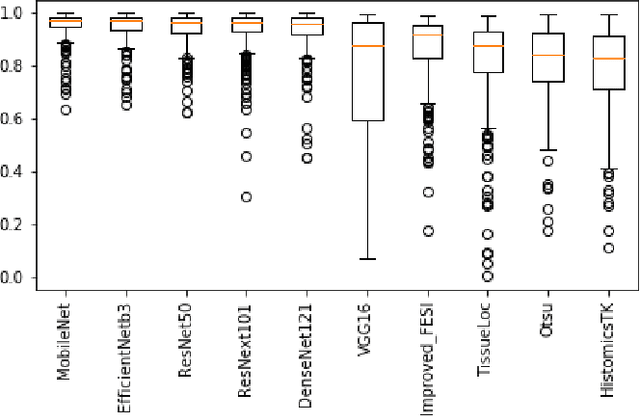
Abstract:During the last decade, the digitization of pathology has gained considerable momentum. Digital pathology offers many advantages including more efficient workflows, easier collaboration as well as a powerful venue for telepathology. At the same time, applying Computer-Aided Diagnosis (CAD) on Whole Slide Images (WSIs) has received substantial attention as a direct result of the digitization. The first step in any image analysis is to extract the tissue. Hence, background removal is an essential prerequisite for efficient and accurate results for many algorithms. In spite of the obvious discrimination for human operators, the identification of tissue regions in WSIs could be challenging for computers, mainly due to the existence of color variations and artifacts. Moreover, some cases such as alveolar tissue types, fatty tissues, and tissues with poor staining are difficult to detect. In this paper, we perform experiments on U-Net architecture with different network backbones (different topologies) to remove the background as well as artifacts from WSIs in order to extract the tissue regions. We compare a wide range of backbone networks including MobileNet, VGG16, EfficientNet-B3, ResNet50, ResNext101 and DenseNet121. We trained and evaluated the network on a manually labeled subset of The Cancer Genome Atlas (TCGA) Dataset. EfficientNet-B3 and MobileNet by almost 99% sensitivity and specificity reached the best results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge