Morteza Babaie
Kimia Lab, University of Waterloo, Waterloo, ON, Canada, Vector Institute, MaRS Centre, Toronto, Canada
On Philomatics and Psychomatics for Combining Philosophy and Psychology with Mathematics
Aug 26, 2023Abstract:We propose the concepts of philomatics and psychomatics as hybrid combinations of philosophy and psychology with mathematics. We explain four motivations for this combination which are fulfilling the desire of analytical philosophy, proposing science of philosophy, justifying mathematical algorithms by philosophy, and abstraction in both philosophy and mathematics. We enumerate various examples for philomatics and psychomatics, some of which are explained in more depth. The first example is the analysis of relation between the context principle, semantic holism, and the usage theory of meaning with the attention mechanism in mathematics. The other example is on the relations of Plato's theory of forms in philosophy with the holographic principle in string theory, object-oriented programming, and machine learning. Finally, the relation between Wittgenstein's family resemblance and clustering in mathematics is explained. This paper opens the door of research for combining philosophy and psychology with mathematics.
An Investigation into Glomeruli Detection in Kidney H&E and PAS Images using YOLO
Jul 25, 2023



Abstract:Context: Analyzing digital pathology images is necessary to draw diagnostic conclusions by investigating tissue patterns and cellular morphology. However, manual evaluation can be time-consuming, expensive, and prone to inter- and intra-observer variability. Objective: To assist pathologists using computerized solutions, automated tissue structure detection and segmentation must be proposed. Furthermore, generating pixel-level object annotations for histopathology images is expensive and time-consuming. As a result, detection models with bounding box labels may be a feasible solution. Design: This paper studies. YOLO-v4 (You-Only-Look-Once), a real-time object detector for microscopic images. YOLO uses a single neural network to predict several bounding boxes and class probabilities for objects of interest. YOLO can enhance detection performance by training on whole slide images. YOLO-v4 has been used in this paper. for glomeruli detection in human kidney images. Multiple experiments have been designed and conducted based on different training data of two public datasets and a private dataset from the University of Michigan for fine-tuning the model. The model was tested on the private dataset from the University of Michigan, serving as an external validation of two different stains, namely hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS). Results: Average specificity and sensitivity for all experiments, and comparison of existing segmentation methods on the same datasets are discussed. Conclusions: Automated glomeruli detection in human kidney images is possible using modern AI models. The design and validation for different stains still depends on variability of public multi-stain datasets.
Immunohistochemistry Biomarkers-Guided Image Search for Histopathology
Apr 24, 2023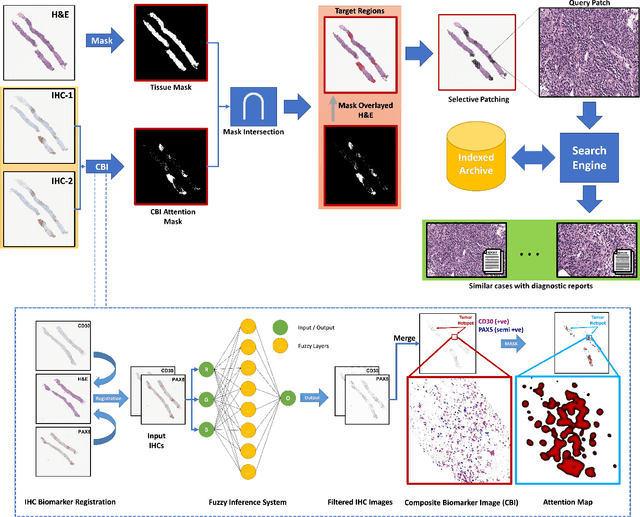
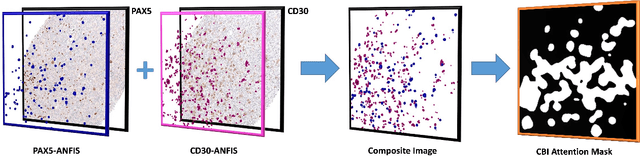
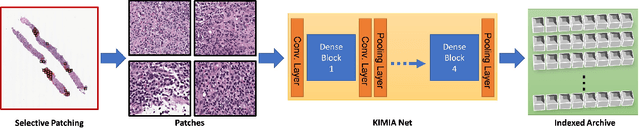
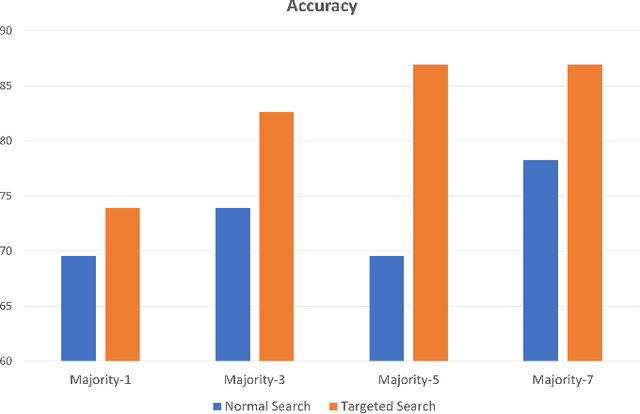
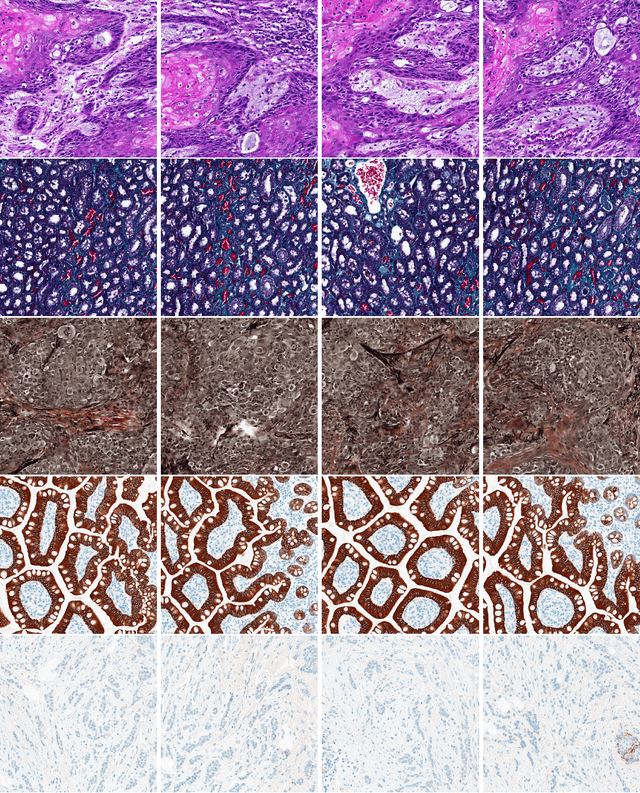
Abstract:Medical practitioners use a number of diagnostic tests to make a reliable diagnosis. Traditionally, Haematoxylin and Eosin (H&E) stained glass slides have been used for cancer diagnosis and tumor detection. However, recently a variety of immunohistochemistry (IHC) stained slides can be requested by pathologists to examine and confirm diagnoses for determining the subtype of a tumor when this is difficult using H&E slides only. Deep learning (DL) has received a lot of interest recently for image search engines to extract features from tissue regions, which may or may not be the target region for diagnosis. This approach generally fails to capture high-level patterns corresponding to the malignant or abnormal content of histopathology images. In this work, we are proposing a targeted image search approach, inspired by the pathologists workflow, which may use information from multiple IHC biomarker images when available. These IHC images could be aligned, filtered, and merged together to generate a composite biomarker image (CBI) that could eventually be used to generate an attention map to guide the search engine for localized search. In our experiments, we observed that an IHC-guided image search engine can retrieve relevant data more accurately than a conventional (i.e., H&E-only) search engine without IHC guidance. Moreover, such engines are also able to accurately conclude the subtypes through majority votes.
Composite Biomarker Image for Advanced Visualization in Histopathology
Apr 24, 2023



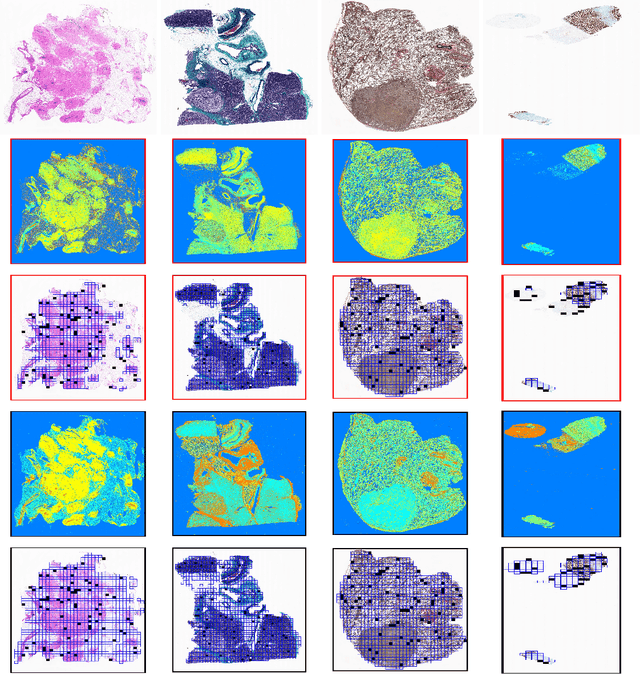
Abstract:Immunohistochemistry (IHC) biomarkers are essential tools for reliable cancer diagnosis and subtyping. It requires cross-staining comparison among Whole Slide Images (WSIs) of IHCs and hematoxylin and eosin (H&E) slides. Currently, pathologists examine the visually co-localized areas across IHC and H&E glass slides for a final diagnosis, which is a tedious and challenging task. Moreover, visually inspecting different IHC slides back and forth to analyze local co-expressions is inherently subjective and prone to error, even when carried out by experienced pathologists. Relying on digital pathology, we propose Composite Biomarker Image (CBI) in this work. CBI is a single image that can be composed using different filtered IHC biomarker images for better visualization. We present a CBI image produced in two steps by the proposed solution for better visualization and hence more efficient clinical workflow. In the first step, IHC biomarker images are aligned with the H&E images using one coordinate system and orientation. In the second step, the positive or negative IHC regions from each biomarker image (based on the pathologists recommendation) are filtered and combined into one image using a fuzzy inference system. For evaluation, the resulting CBI images, from the proposed system, were evaluated qualitatively by the expert pathologists. The CBI concept helps the pathologists to identify the suspected target tissues more easily, which could be further assessed by examining the actual WSIs at the same suspected regions.
Learning Binary and Sparse Permutation-Invariant Representations for Fast and Memory Efficient Whole Slide Image Search
Aug 29, 2022


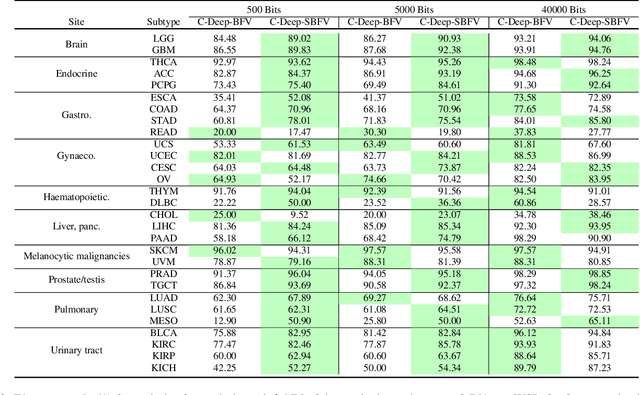
Abstract:Learning suitable Whole slide images (WSIs) representations for efficient retrieval systems is a non-trivial task. The WSI embeddings obtained from current methods are in Euclidean space not ideal for efficient WSI retrieval. Furthermore, most of the current methods require high GPU memory due to the simultaneous processing of multiple sets of patches. To address these challenges, we propose a novel framework for learning binary and sparse WSI representations utilizing a deep generative modelling and the Fisher Vector. We introduce new loss functions for learning sparse and binary permutation-invariant WSI representations that employ instance-based training achieving better memory efficiency. The learned WSI representations are validated on The Cancer Genomic Atlas (TCGA) and Liver-Kidney-Stomach (LKS) datasets. The proposed method outperforms Yottixel (a recent search engine for histopathology images) both in terms of retrieval accuracy and speed. Further, we achieve competitive performance against SOTA on the public benchmark LKS dataset for WSI classification.
Automatic Multi-Stain Registration of Whole Slide Images in Histopathology
Jul 29, 2021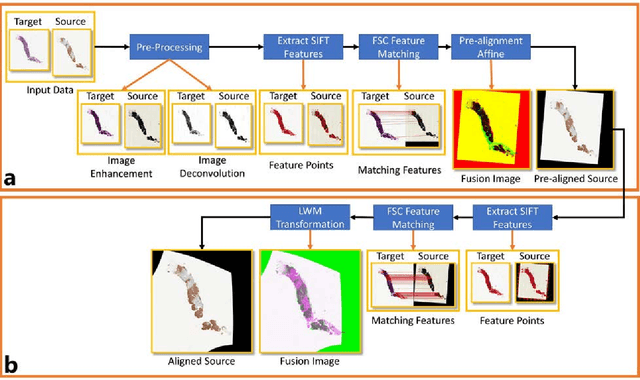
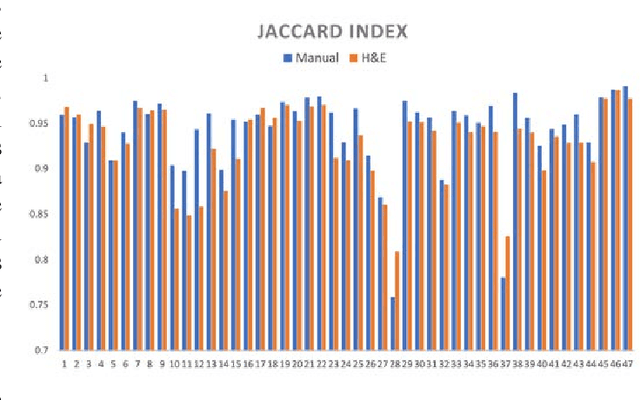
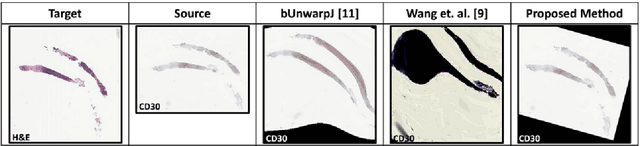
Abstract:Joint analysis of multiple biomarker images and tissue morphology is important for disease diagnosis, treatment planning and drug development. It requires cross-staining comparison among Whole Slide Images (WSIs) of immuno-histochemical and hematoxylin and eosin (H&E) microscopic slides. However, automatic, and fast cross-staining alignment of enormous gigapixel WSIs at single-cell precision is challenging. In addition to morphological deformations introduced during slide preparation, there are large variations in cell appearance and tissue morphology across different staining. In this paper, we propose a two-step automatic feature-based cross-staining WSI alignment to assist localization of even tiny metastatic foci in the assessment of lymph node. Image pairs were aligned allowing for translation, rotation, and scaling. The registration was performed automatically by first detecting landmarks in both images, using the scale-invariant image transform (SIFT), followed by the fast sample consensus (FSC) protocol for finding point correspondences and finally aligned the images. The Registration results were evaluated using both visual and quantitative criteria using the Jaccard index. The average Jaccard similarity index of the results produced by the proposed system is 0.942 when compared with the manual registration.
Colored Kimia Path24 Dataset: Configurations and Benchmarks with Deep Embeddings
Feb 15, 2021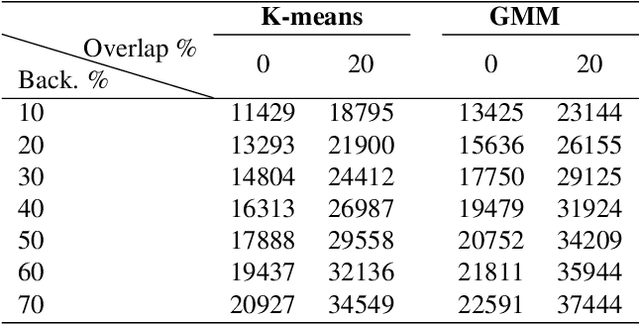
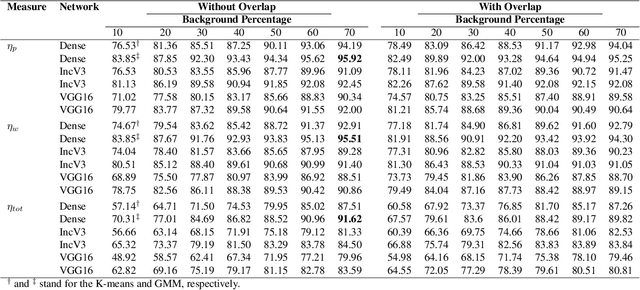
Abstract:The Kimia Path24 dataset has been introduced as a classification and retrieval dataset for digital pathology. Although it provides multi-class data, the color information has been neglected in the process of extracting patches. The staining information plays a major role in the recognition of tissue patterns. To address this drawback, we introduce the color version of Kimia Path24 by recreating sample patches from all 24 scans to propose Kimia Path24C. We run extensive experiments to determine the best configuration for selected patches. To provide preliminary results for setting a benchmark for the new dataset, we utilize VGG16, InceptionV3 and DenseNet-121 model as feature extractors. Then, we use these feature vectors to retrieve test patches. The accuracy of image retrieval using DenseNet was 95.92% while the highest accuracy using InceptionV3 and VGG16 reached 92.45% and 92%, respectively. We also experimented with "deep barcodes" and established that with a small loss in accuracy (e.g., 93.43% for binarized features for DenseNet instead of 95.92% when the features themselves are used), the search operations can be significantly accelerated.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021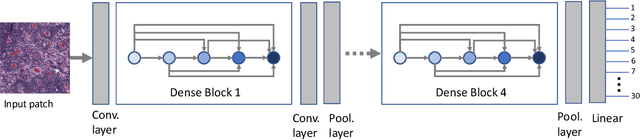
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Ink Marker Segmentation in Histopathology Images Using Deep Learning
Oct 29, 2020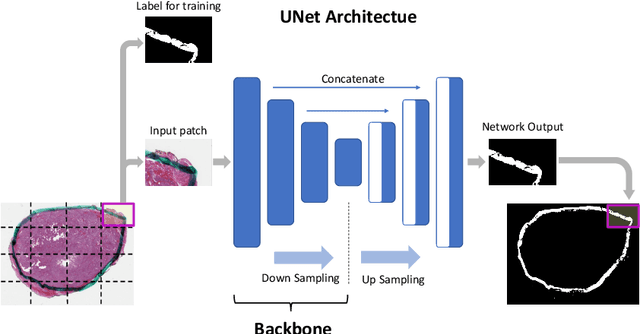
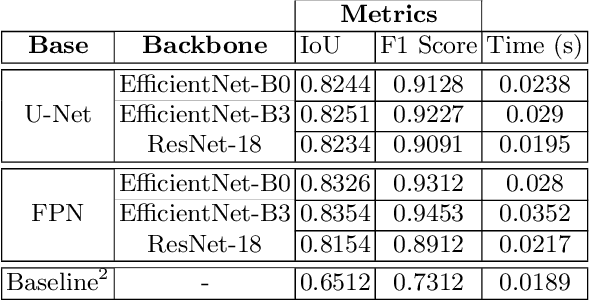
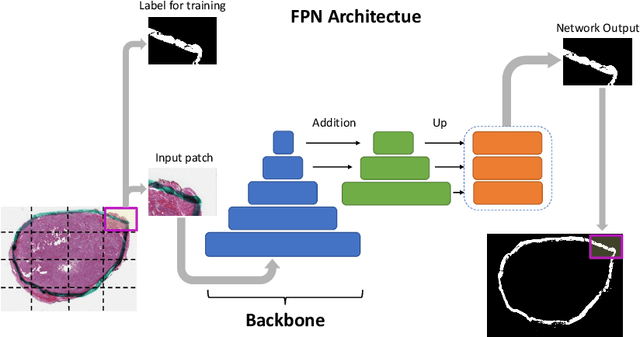
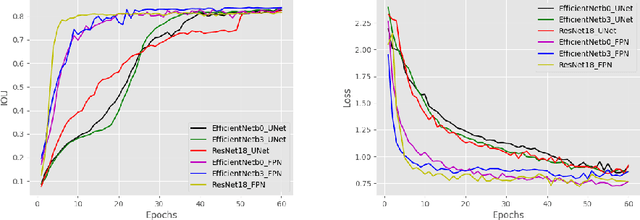
Abstract:Due to the recent advancements in machine vision, digital pathology has gained significant attention. Histopathology images are distinctly rich in visual information. The tissue glass slide images are utilized for disease diagnosis. Researchers study many methods to process histopathology images and facilitate fast and reliable diagnosis; therefore, the availability of high-quality slides becomes paramount. The quality of the images can be negatively affected when the glass slides are ink-marked by pathologists to delineate regions of interest. As an example, in one of the largest public histopathology datasets, The Cancer Genome Atlas (TCGA), approximately $12\%$ of the digitized slides are affected by manual delineations through ink markings. To process these open-access slide images and other repositories for the design and validation of new methods, an algorithm to detect the marked regions of the images is essential to avoid confusing tissue pixels with ink-colored pixels for computer methods. In this study, we propose to segment the ink-marked areas of pathology patches through a deep network. A dataset from $79$ whole slide images with $4,305$ patches was created and different networks were trained. Finally, the results showed an FPN model with the EffiecentNet-B3 as the backbone was found to be the superior configuration with an F1 score of $94.53\%$.
Forming Local Intersections of Projections for Classifying and Searching Histopathology Images
Aug 08, 2020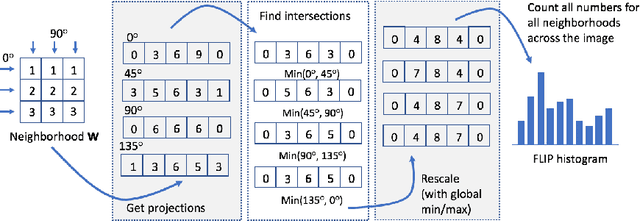
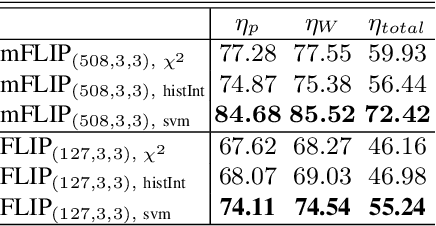
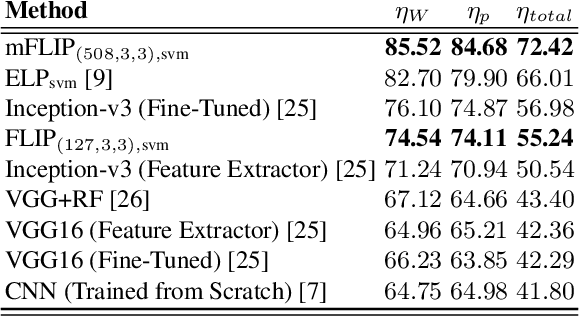

Abstract:In this paper, we propose a novel image descriptor called Forming Local Intersections of Projections (FLIP) and its multi-resolution version (mFLIP) for representing histopathology images. The descriptor is based on the Radon transform wherein we apply parallel projections in small local neighborhoods of gray-level images. Using equidistant projection directions in each window, we extract unique and invariant characteristics of the neighborhood by taking the intersection of adjacent projections. Thereafter, we construct a histogram for each image, which we call the FLIP histogram. Various resolutions provide different FLIP histograms which are then concatenated to form the mFLIP descriptor. Our experiments included training common networks from scratch and fine-tuning pre-trained networks to benchmark our proposed descriptor. Experiments are conducted on the publicly available dataset KIMIA Path24 and KIMIA Path960. For both of these datasets, FLIP and mFLIP descriptors show promising results in all experiments.Using KIMIA Path24 data, FLIP outperformed non-fine-tuned Inception-v3 and fine-tuned VGG16 and mFLIP outperformed fine-tuned Inception-v3 in feature extracting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge