Shivam Kalra
Structured Model Pruning for Efficient Inference in Computational Pathology
Apr 12, 2024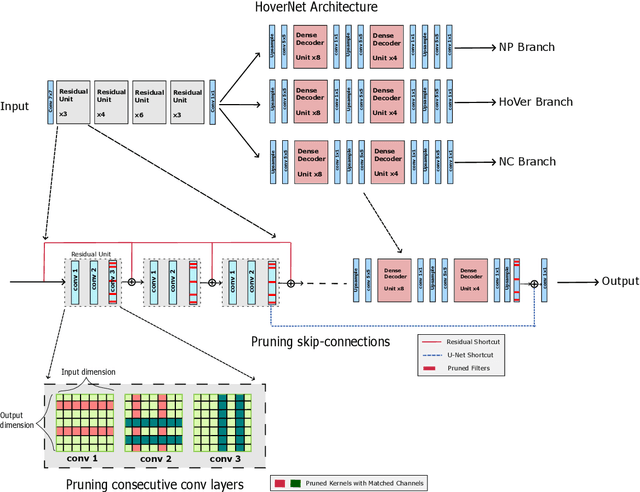

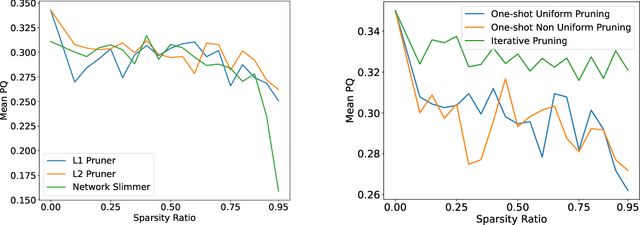

Abstract:Recent years have seen significant efforts to adopt Artificial Intelligence (AI) in healthcare for various use cases, from computer-aided diagnosis to ICU triage. However, the size of AI models has been rapidly growing due to scaling laws and the success of foundational models, which poses an increasing challenge to leverage advanced models in practical applications. It is thus imperative to develop efficient models, especially for deploying AI solutions under resource-constrains or with time sensitivity. One potential solution is to perform model compression, a set of techniques that remove less important model components or reduce parameter precision, to reduce model computation demand. In this work, we demonstrate that model pruning, as a model compression technique, can effectively reduce inference cost for computational and digital pathology based analysis with a negligible loss of analysis performance. To this end, we develop a methodology for pruning the widely used U-Net-style architectures in biomedical imaging, with which we evaluate multiple pruning heuristics on nuclei instance segmentation and classification, and empirically demonstrate that pruning can compress models by at least 70% with a negligible drop in performance.
Comments on 'Fast and scalable search of whole-slide images via self-supervised deep learning'
Apr 18, 2023Abstract:Chen et al. [Chen2022] recently published the article 'Fast and scalable search of whole-slide images via self-supervised deep learning' in Nature Biomedical Engineering. The authors call their method 'self-supervised image search for histology', short SISH. We express our concerns that SISH is an incremental modification of Yottixel, has used MinMax binarization but does not cite the original works, and is based on a misnomer 'self-supervised image search'. As well, we point to several other concerns regarding experiments and comparisons performed by Chen et al.
Learning Binary and Sparse Permutation-Invariant Representations for Fast and Memory Efficient Whole Slide Image Search
Aug 29, 2022


Abstract:Learning suitable Whole slide images (WSIs) representations for efficient retrieval systems is a non-trivial task. The WSI embeddings obtained from current methods are in Euclidean space not ideal for efficient WSI retrieval. Furthermore, most of the current methods require high GPU memory due to the simultaneous processing of multiple sets of patches. To address these challenges, we propose a novel framework for learning binary and sparse WSI representations utilizing a deep generative modelling and the Fisher Vector. We introduce new loss functions for learning sparse and binary permutation-invariant WSI representations that employ instance-based training achieving better memory efficiency. The learned WSI representations are validated on The Cancer Genomic Atlas (TCGA) and Liver-Kidney-Stomach (LKS) datasets. The proposed method outperforms Yottixel (a recent search engine for histopathology images) both in terms of retrieval accuracy and speed. Further, we achieve competitive performance against SOTA on the public benchmark LKS dataset for WSI classification.
ProxyFL: Decentralized Federated Learning through Proxy Model Sharing
Nov 22, 2021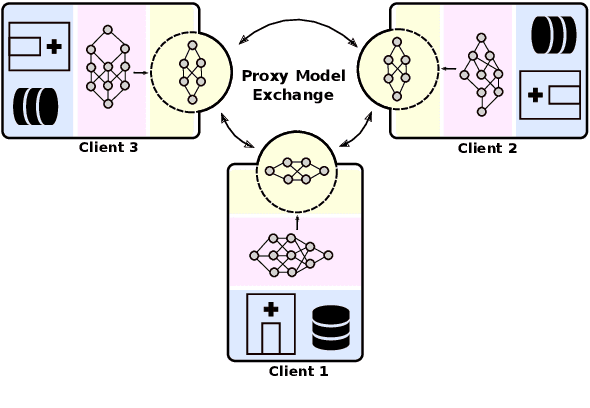
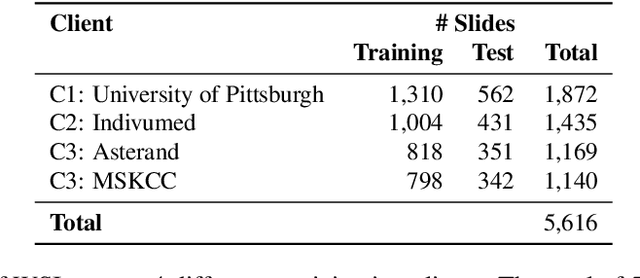
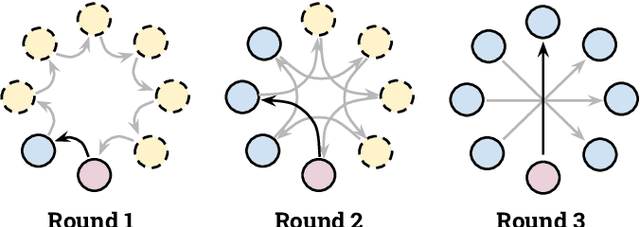
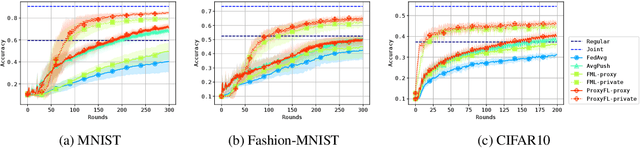
Abstract:Institutions in highly regulated domains such as finance and healthcare often have restrictive rules around data sharing. Federated learning is a distributed learning framework that enables multi-institutional collaborations on decentralized data with improved protection for each collaborator's data privacy. In this paper, we propose a communication-efficient scheme for decentralized federated learning called ProxyFL, or proxy-based federated learning. Each participant in ProxyFL maintains two models, a private model, and a publicly shared proxy model designed to protect the participant's privacy. Proxy models allow efficient information exchange among participants using the PushSum method without the need of a centralized server. The proposed method eliminates a significant limitation of canonical federated learning by allowing model heterogeneity; each participant can have a private model with any architecture. Furthermore, our protocol for communication by proxy leads to stronger privacy guarantees using differential privacy analysis. Experiments on popular image datasets, and a pan-cancer diagnostic problem using over 30,000 high-quality gigapixel histology whole slide images, show that ProxyFL can outperform existing alternatives with much less communication overhead and stronger privacy.
Pay Attention with Focus: A Novel Learning Scheme for Classification of Whole Slide Images
Jun 11, 2021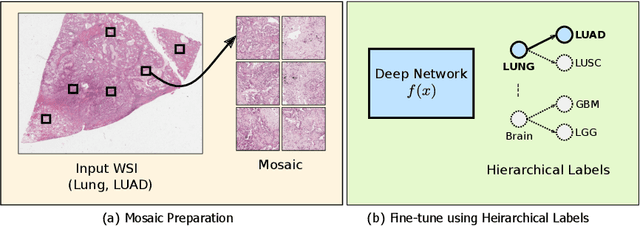
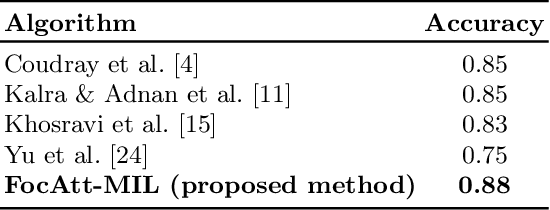
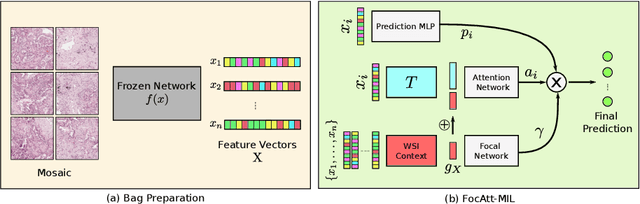
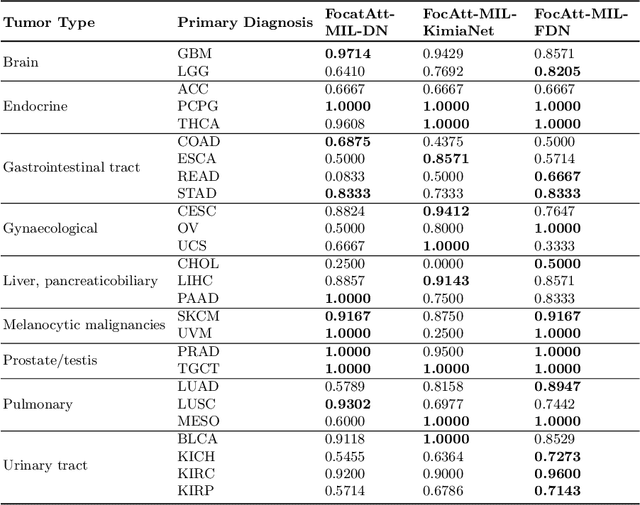
Abstract:Deep learning methods such as convolutional neural networks (CNNs) are difficult to directly utilize to analyze whole slide images (WSIs) due to the large image dimensions. We overcome this limitation by proposing a novel two-stage approach. First, we extract a set of representative patches (called mosaic) from a WSI. Each patch of a mosaic is encoded to a feature vector using a deep network. The feature extractor model is fine-tuned using hierarchical target labels of WSIs, i.e., anatomic site and primary diagnosis. In the second stage, a set of encoded patch-level features from a WSI is used to compute the primary diagnosis probability through the proposed Pay Attention with Focus scheme, an attention-weighted averaging of predicted probabilities for all patches of a mosaic modulated by a trainable focal factor. Experimental results show that the proposed model can be robust, and effective for the classification of WSIs.
Colored Kimia Path24 Dataset: Configurations and Benchmarks with Deep Embeddings
Feb 15, 2021
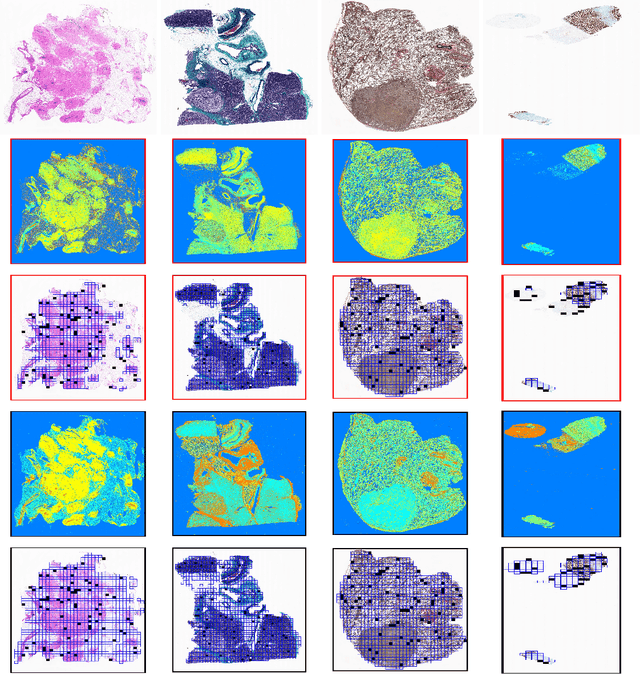
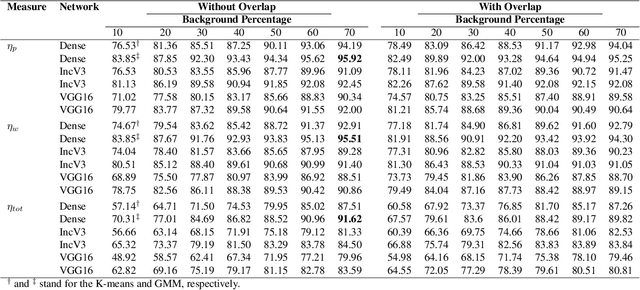
Abstract:The Kimia Path24 dataset has been introduced as a classification and retrieval dataset for digital pathology. Although it provides multi-class data, the color information has been neglected in the process of extracting patches. The staining information plays a major role in the recognition of tissue patterns. To address this drawback, we introduce the color version of Kimia Path24 by recreating sample patches from all 24 scans to propose Kimia Path24C. We run extensive experiments to determine the best configuration for selected patches. To provide preliminary results for setting a benchmark for the new dataset, we utilize VGG16, InceptionV3 and DenseNet-121 model as feature extractors. Then, we use these feature vectors to retrieve test patches. The accuracy of image retrieval using DenseNet was 95.92% while the highest accuracy using InceptionV3 and VGG16 reached 92.45% and 92%, respectively. We also experimented with "deep barcodes" and established that with a small loss in accuracy (e.g., 93.43% for binarized features for DenseNet instead of 95.92% when the features themselves are used), the search operations can be significantly accelerated.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021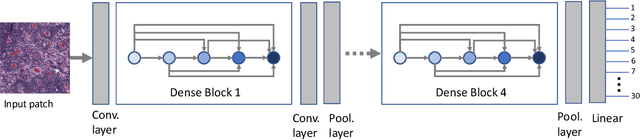
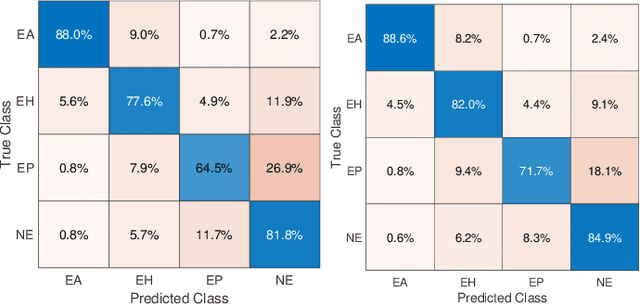
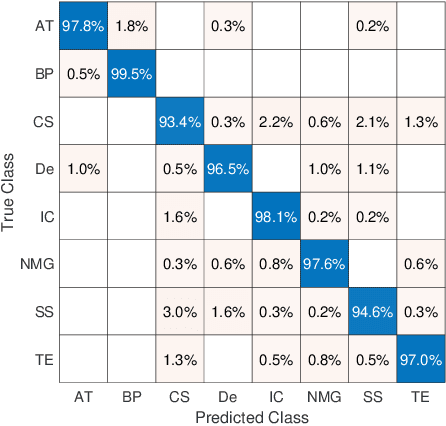
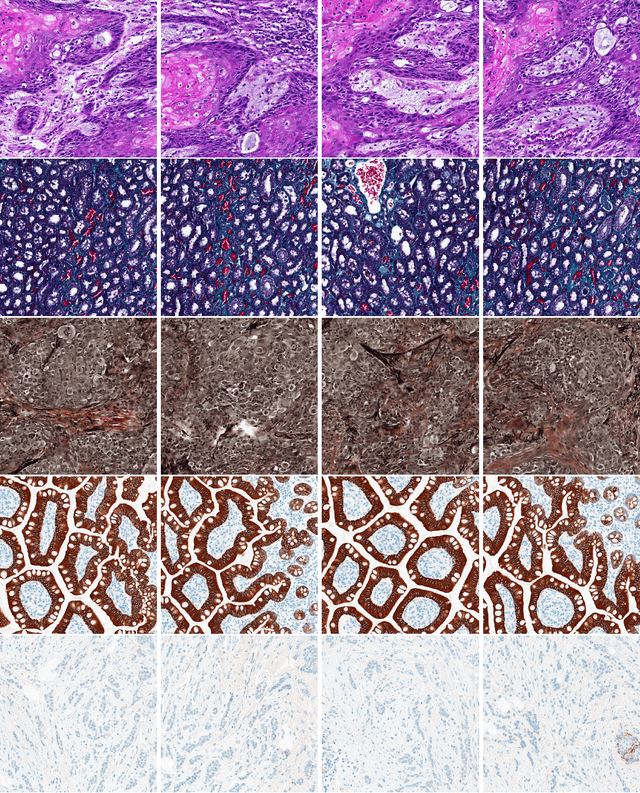
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Forming Local Intersections of Projections for Classifying and Searching Histopathology Images
Aug 08, 2020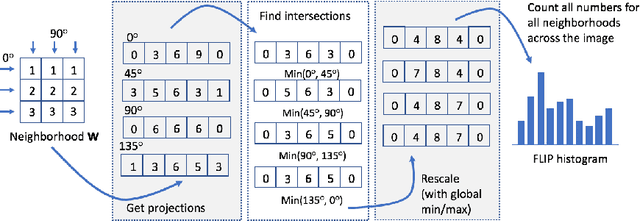
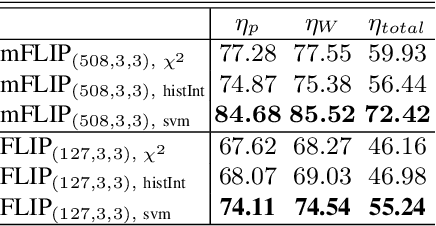
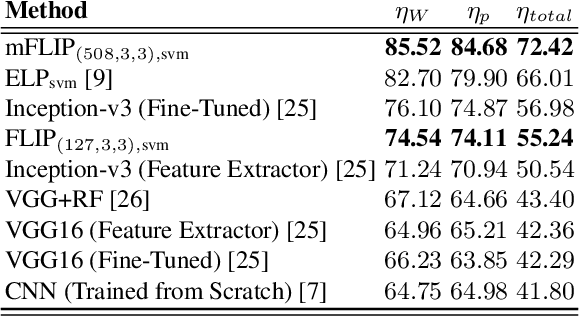

Abstract:In this paper, we propose a novel image descriptor called Forming Local Intersections of Projections (FLIP) and its multi-resolution version (mFLIP) for representing histopathology images. The descriptor is based on the Radon transform wherein we apply parallel projections in small local neighborhoods of gray-level images. Using equidistant projection directions in each window, we extract unique and invariant characteristics of the neighborhood by taking the intersection of adjacent projections. Thereafter, we construct a histogram for each image, which we call the FLIP histogram. Various resolutions provide different FLIP histograms which are then concatenated to form the mFLIP descriptor. Our experiments included training common networks from scratch and fine-tuning pre-trained networks to benchmark our proposed descriptor. Experiments are conducted on the publicly available dataset KIMIA Path24 and KIMIA Path960. For both of these datasets, FLIP and mFLIP descriptors show promising results in all experiments.Using KIMIA Path24 data, FLIP outperformed non-fine-tuned Inception-v3 and fine-tuned VGG16 and mFLIP outperformed fine-tuned Inception-v3 in feature extracting.
Recognizing Magnification Levels in Microscopic Snapshots
May 07, 2020


Abstract:Recent advances in digital imaging has transformed computer vision and machine learning to new tools for analyzing pathology images. This trend could automate some of the tasks in the diagnostic pathology and elevate the pathologist workload. The final step of any cancer diagnosis procedure is performed by the expert pathologist. These experts use microscopes with high level of optical magnification to observe minute characteristics of the tissue acquired through biopsy and fixed on glass slides. Switching between different magnifications, and finding the magnification level at which they identify the presence or absence of malignant tissues is important. As the majority of pathologists still use light microscopy, compared to digital scanners, in many instance a mounted camera on the microscope is used to capture snapshots from significant field-of-views. Repositories of such snapshots usually do not contain the magnification information. In this paper, we extract deep features of the images available on TCGA dataset with known magnification to train a classifier for magnification recognition. We compared the results with LBP, a well-known handcrafted feature extraction method. The proposed approach achieved a mean accuracy of 96% when a multi-layer perceptron was trained as a classifier.
Representation Learning of Histopathology Images using Graph Neural Networks
Apr 17, 2020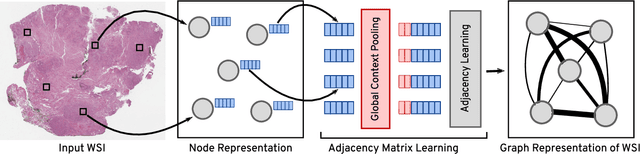
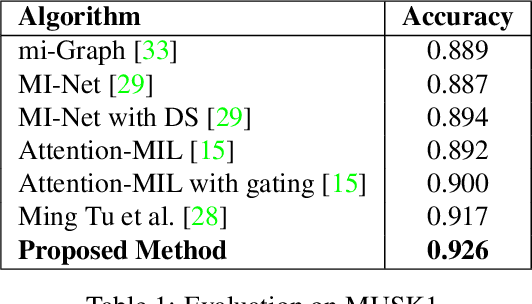
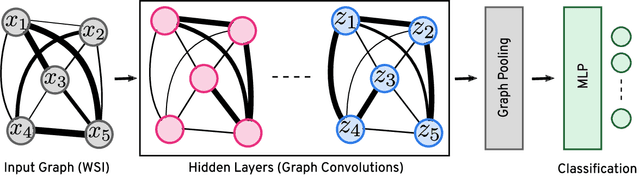
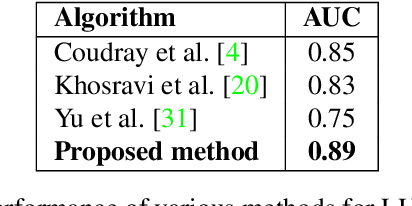
Abstract:Representation learning for Whole Slide Images (WSIs) is pivotal in developing image-based systems to achieve higher precision in diagnostic pathology. We propose a two-stage framework for WSI representation learning. We sample relevant patches using a color-based method and use graph neural networks to learn relations among sampled patches to aggregate the image information into a single vector representation. We introduce attention via graph pooling to automatically infer patches with higher relevance. We demonstrate the performance of our approach for discriminating two sub-types of lung cancers, Lung Adenocarcinoma (LUAD) & Lung Squamous Cell Carcinoma (LUSC). We collected 1,026 lung cancer WSIs with the 40$\times$ magnification from The Cancer Genome Atlas (TCGA) dataset, the largest public repository of histopathology images and achieved state-of-the-art accuracy of 88.8% and AUC of 0.89 on lung cancer sub-type classification by extracting features from a pre-trained DenseNet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge