Nazim Shaikh
When RAG Hurts: Diagnosing and Mitigating Attention Distraction in Retrieval-Augmented LVLMs
Jan 30, 2026Abstract:While Retrieval-Augmented Generation (RAG) is one of the dominant paradigms for enhancing Large Vision-Language Models (LVLMs) on knowledge-based VQA tasks, recent work attributes RAG failures to insufficient attention towards the retrieved context, proposing to reduce the attention allocated to image tokens. In this work, we identify a distinct failure mode that previous study overlooked: Attention Distraction (AD). When the retrieved context is sufficient (highly relevant or including the correct answer), the retrieved text suppresses the visual attention globally, and the attention on image tokens shifts away from question-relevant regions. This leads to failures on questions the model could originally answer correctly without the retrieved text. To mitigate this issue, we propose MAD-RAG, a training-free intervention that decouples visual grounding from context integration through a dual-question formulation, combined with attention mixing to preserve image-conditioned evidence. Extensive experiments on OK-VQA, E-VQA, and InfoSeek demonstrate that MAD-RAG consistently outperforms existing baselines across different model families, yielding absolute gains of up to 4.76%, 9.20%, and 6.18% over the vanilla RAG baseline. Notably, MAD-RAG rectifies up to 74.68% of failure cases with negligible computational overhead.
Structured Model Pruning for Efficient Inference in Computational Pathology
Apr 12, 2024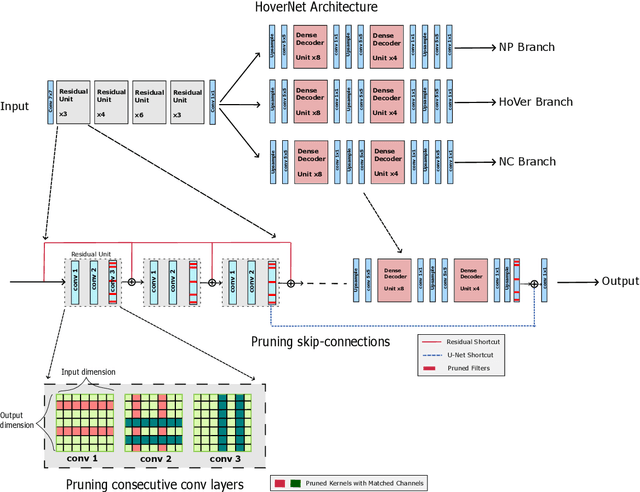

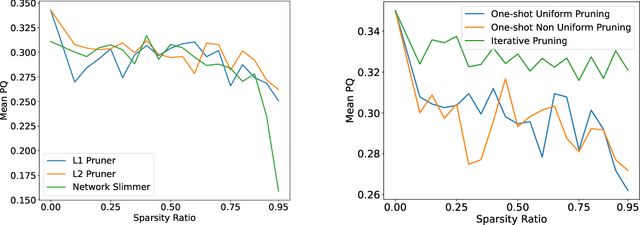

Abstract:Recent years have seen significant efforts to adopt Artificial Intelligence (AI) in healthcare for various use cases, from computer-aided diagnosis to ICU triage. However, the size of AI models has been rapidly growing due to scaling laws and the success of foundational models, which poses an increasing challenge to leverage advanced models in practical applications. It is thus imperative to develop efficient models, especially for deploying AI solutions under resource-constrains or with time sensitivity. One potential solution is to perform model compression, a set of techniques that remove less important model components or reduce parameter precision, to reduce model computation demand. In this work, we demonstrate that model pruning, as a model compression technique, can effectively reduce inference cost for computational and digital pathology based analysis with a negligible loss of analysis performance. To this end, we develop a methodology for pruning the widely used U-Net-style architectures in biomedical imaging, with which we evaluate multiple pruning heuristics on nuclei instance segmentation and classification, and empirically demonstrate that pruning can compress models by at least 70% with a negligible drop in performance.
Cross-modality Attention-based Multimodal Fusion for Non-small Cell Lung Cancer (NSCLC) Patient Survival Prediction
Aug 18, 2023Abstract:Cancer prognosis and survival outcome predictions are crucial for therapeutic response estimation and for stratifying patients into various treatment groups. Medical domains concerned with cancer prognosis are abundant with multiple modalities, including pathological image data and non-image data such as genomic information. To date, multimodal learning has shown potential to enhance clinical prediction model performance by extracting and aggregating information from different modalities of the same subject. This approach could outperform single modality learning, thus improving computer-aided diagnosis and prognosis in numerous medical applications. In this work, we propose a cross-modality attention-based multimodal fusion pipeline designed to integrate modality-specific knowledge for patient survival prediction in non-small cell lung cancer (NSCLC). Instead of merely concatenating or summing up the features from different modalities, our method gauges the importance of each modality for feature fusion with cross-modality relationship when infusing the multimodal features. Compared with single modality, which achieved c-index of 0.5772 and 0.5885 using solely tissue image data or RNA-seq data, respectively, the proposed fusion approach achieved c-index 0.6587 in our experiment, showcasing the capability of assimilating modality-specific knowledge from varied modalities.
Why Should we Combine Training and Post-Training Methods for Out-of-Distribution Detection?
Dec 05, 2019


Abstract:Deep neural networks are known to achieve superior results in classification tasks. However, it has been recently shown that they are incapable to detect examples that are generated by a distribution which is different than the one they have been trained on since they are making overconfident prediction for Out-Of-Distribution (OOD) examples. OOD detection has attracted a lot of attention recently. In this paper, we review some of the most seminal recent algorithms in the OOD detection field, we divide those methods into training and post-training and we experimentally show how the combination of the former with the latter can achieve state-of-the-art results in the OOD detection task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge