Yao Nie
Visible Light Positioning With Lamé Curve LEDs: A Generic Approach for Camera Pose Estimation
Feb 02, 2026Abstract:Camera-based visible light positioning (VLP) is a promising technique for accurate and low-cost indoor camera pose estimation (CPE). To reduce the number of required light-emitting diodes (LEDs), advanced methods commonly exploit LED shape features for positioning. Although interesting, they are typically restricted to a single LED geometry, leading to failure in heterogeneous LED-shape scenarios. To address this challenge, this paper investigates Lamé curves as a unified representation of common LED shapes and proposes a generic VLP algorithm using Lamé curve-shaped LEDs, termed LC-VLP. In the considered system, multiple ceiling-mounted Lamé curve-shaped LEDs periodically broadcast their curve parameters via visible light communication, which are captured by a camera-equipped receiver. Based on the received LED images and curve parameters, the receiver can estimate the camera pose using LC-VLP. Specifically, an LED database is constructed offline to store the curve parameters, while online positioning is formulated as a nonlinear least-squares problem and solved iteratively. To provide a reliable initialization, a correspondence-free perspective-\textit{n}-points (FreeP\textit{n}P) algorithm is further developed, enabling approximate CPE without any pre-calibrated reference points. The performance of LC-VLP is verified by both simulations and experiments. Simulations show that LC-VLP outperforms state-of-the-art methods in both circular- and rectangular-LED scenarios, achieving reductions of over 40% in position error and 25% in rotation error. Experiments further show that LC-VLP can achieve an average position accuracy of less than 4 cm.
When RAG Hurts: Diagnosing and Mitigating Attention Distraction in Retrieval-Augmented LVLMs
Jan 30, 2026Abstract:While Retrieval-Augmented Generation (RAG) is one of the dominant paradigms for enhancing Large Vision-Language Models (LVLMs) on knowledge-based VQA tasks, recent work attributes RAG failures to insufficient attention towards the retrieved context, proposing to reduce the attention allocated to image tokens. In this work, we identify a distinct failure mode that previous study overlooked: Attention Distraction (AD). When the retrieved context is sufficient (highly relevant or including the correct answer), the retrieved text suppresses the visual attention globally, and the attention on image tokens shifts away from question-relevant regions. This leads to failures on questions the model could originally answer correctly without the retrieved text. To mitigate this issue, we propose MAD-RAG, a training-free intervention that decouples visual grounding from context integration through a dual-question formulation, combined with attention mixing to preserve image-conditioned evidence. Extensive experiments on OK-VQA, E-VQA, and InfoSeek demonstrate that MAD-RAG consistently outperforms existing baselines across different model families, yielding absolute gains of up to 4.76%, 9.20%, and 6.18% over the vanilla RAG baseline. Notably, MAD-RAG rectifies up to 74.68% of failure cases with negligible computational overhead.
A Novel Generative Artificial Intelligence Method for Interference Study on Multiplex Brightfield Immunohistochemistry Images
Aug 15, 2024



Abstract:Multiplex brightfield imaging offers the advantage of simultaneously analyzing multiple biomarkers on a single slide, as opposed to single biomarker labeling on multiple consecutive slides. To accurately analyze multiple biomarkers localized at the same cellular compartment, two representative biomarker sets were selected as assay models - cMET-PDL1-EGFR and CD8-LAG3-PDL1, where all three biomarkers can co-localize on the cell membrane. One of the most crucial preliminary stages for analyzing such assay is identifying each unique chromogen on individual cells. This is a challenging problem due to the co-localization of membrane stains from all the three biomarkers. It requires advanced color unmixing for creating the equivalent singleplex images from each triplex image for each biomarker. In this project, we developed a cycle-Generative Adversarial Network (cycle-GAN) method for unmixing the triplex images generated from the above-mentioned assays. Three different models were designed to generate the singleplex image for each of the three stains Tamra (purple), QM-Dabsyl (yellow) and Green. A notable novelty of our approach was that the input to the network were images in the optical density domain instead of conventionally used RGB images. The use of the optical density domain helped in reducing the blurriness of the synthetic singleplex images, which was often observed when the network was trained on RGB images. The cycle-GAN models were validated on 10,800 lung, gastric and colon images for the cMET-PDL1-EGFR assay and 3600 colon images for the CD8-LAG3-PDL1 assay. Visual as well as quantified assessments demonstrated that the proposed method is effective and efficient when compared with the manual reviewing results and is readily applicable to various multiplex assays.
Cross-modality Attention-based Multimodal Fusion for Non-small Cell Lung Cancer (NSCLC) Patient Survival Prediction
Aug 18, 2023Abstract:Cancer prognosis and survival outcome predictions are crucial for therapeutic response estimation and for stratifying patients into various treatment groups. Medical domains concerned with cancer prognosis are abundant with multiple modalities, including pathological image data and non-image data such as genomic information. To date, multimodal learning has shown potential to enhance clinical prediction model performance by extracting and aggregating information from different modalities of the same subject. This approach could outperform single modality learning, thus improving computer-aided diagnosis and prognosis in numerous medical applications. In this work, we propose a cross-modality attention-based multimodal fusion pipeline designed to integrate modality-specific knowledge for patient survival prediction in non-small cell lung cancer (NSCLC). Instead of merely concatenating or summing up the features from different modalities, our method gauges the importance of each modality for feature fusion with cross-modality relationship when infusing the multimodal features. Compared with single modality, which achieved c-index of 0.5772 and 0.5885 using solely tissue image data or RNA-seq data, respectively, the proposed fusion approach achieved c-index 0.6587 in our experiment, showcasing the capability of assimilating modality-specific knowledge from varied modalities.
Weakly Supervised Multi-Task Learning for Cell Detection and Segmentation
Oct 27, 2019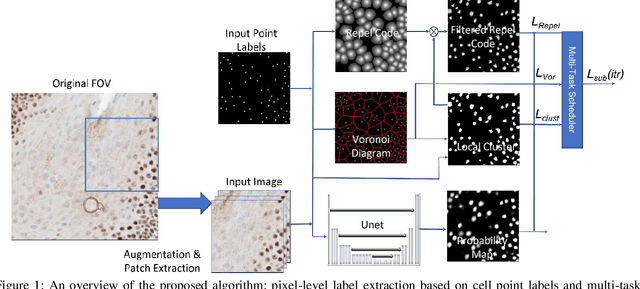
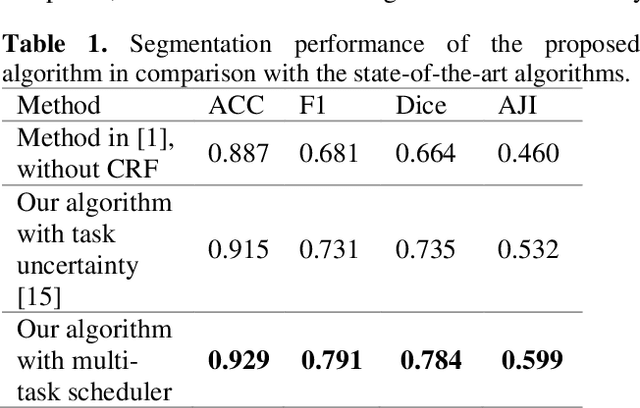
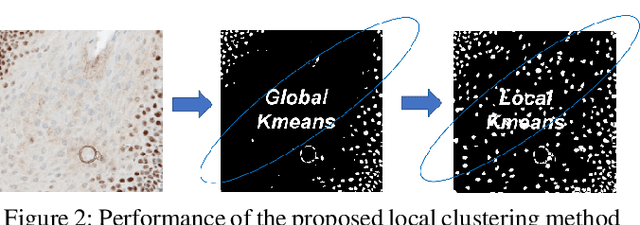
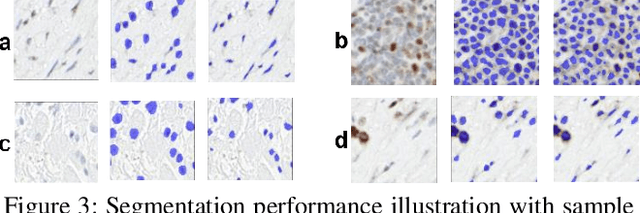
Abstract:Cell detection and segmentation is fundamental for all downstream analysis of digital pathology images. However, obtaining the pixel-level ground truth for single cell segmentation is extremely labor intensive. To overcome this challenge, we developed an end-to-end deep learning algorithm to perform both single cell detection and segmentation using only point labels. This is achieved through the combination of different task orientated point label encoding methods and a multi-task scheduler for training. We apply and validate our algorithm on PMS2 stained colon rectal cancer and tonsil tissue images. Compared to the state-of-the-art, our algorithm shows significant improvement in cell detection and segmentation without increasing the annotation efforts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge