Mohammed Adnan
Sparse Training from Random Initialization: Aligning Lottery Ticket Masks using Weight Symmetry
May 08, 2025Abstract:The Lottery Ticket Hypothesis (LTH) suggests there exists a sparse LTH mask and weights that achieve the same generalization performance as the dense model while using significantly fewer parameters. However, finding a LTH solution is computationally expensive, and a LTH sparsity mask does not generalize to other random weight initializations. Recent work has suggested that neural networks trained from random initialization find solutions within the same basin modulo permutation, and proposes a method to align trained models within the same loss basin. We hypothesize that misalignment of basins is the reason why LTH masks do not generalize to new random initializations and propose permuting the LTH mask to align with the new optimization basin when performing sparse training from a different random init. We empirically show a significant increase in generalization when sparse training from random initialization with the permuted mask as compared to using the non-permuted LTH mask, on multiple datasets (CIFAR-10, CIFAR-100 and ImageNet) and models (VGG11, ResNet20 and ResNet50).
Structured Model Pruning for Efficient Inference in Computational Pathology
Apr 12, 2024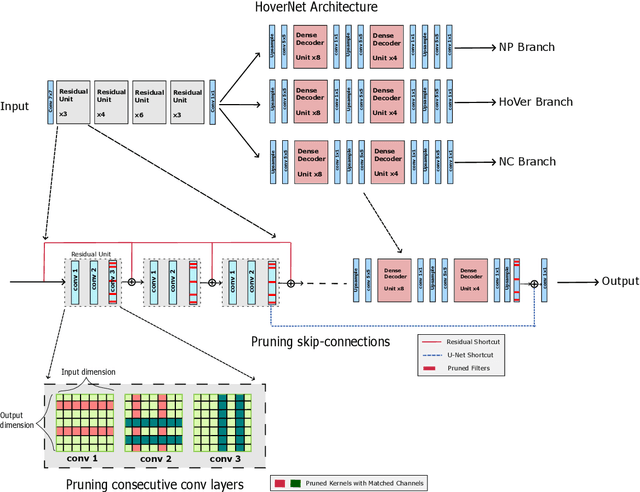

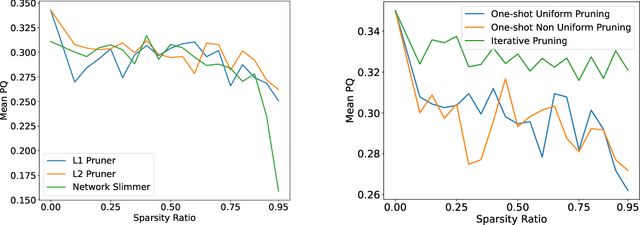

Abstract:Recent years have seen significant efforts to adopt Artificial Intelligence (AI) in healthcare for various use cases, from computer-aided diagnosis to ICU triage. However, the size of AI models has been rapidly growing due to scaling laws and the success of foundational models, which poses an increasing challenge to leverage advanced models in practical applications. It is thus imperative to develop efficient models, especially for deploying AI solutions under resource-constrains or with time sensitivity. One potential solution is to perform model compression, a set of techniques that remove less important model components or reduce parameter precision, to reduce model computation demand. In this work, we demonstrate that model pruning, as a model compression technique, can effectively reduce inference cost for computational and digital pathology based analysis with a negligible loss of analysis performance. To this end, we develop a methodology for pruning the widely used U-Net-style architectures in biomedical imaging, with which we evaluate multiple pruning heuristics on nuclei instance segmentation and classification, and empirically demonstrate that pruning can compress models by at least 70% with a negligible drop in performance.
Monitoring Shortcut Learning using Mutual Information
Jun 27, 2022
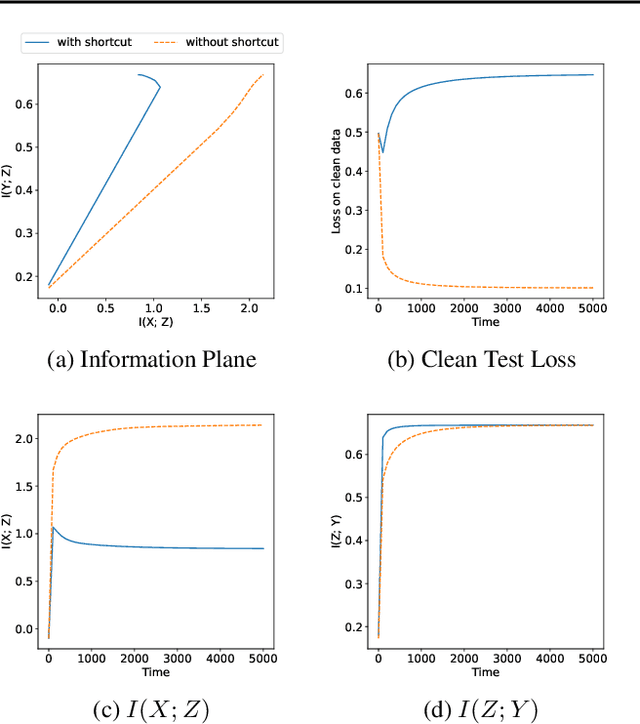
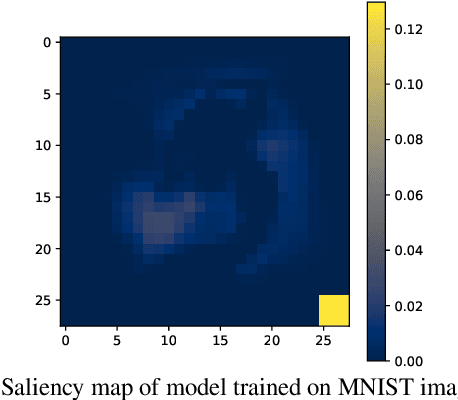
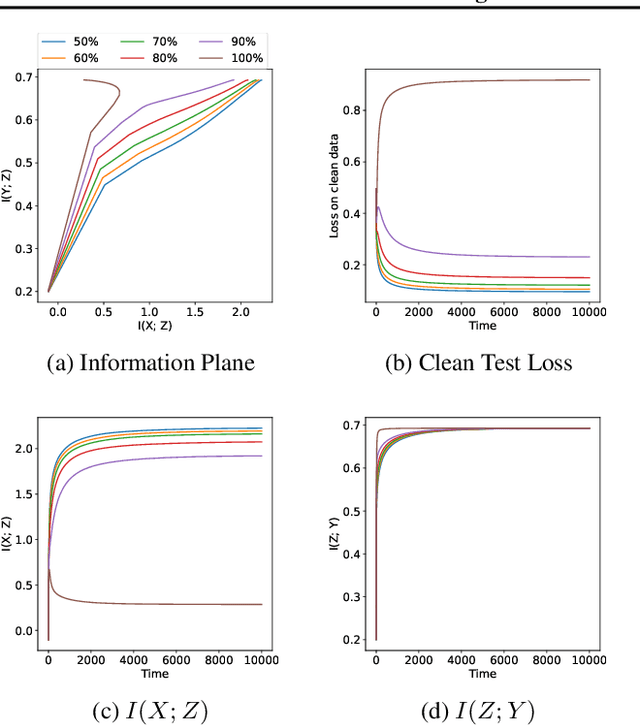
Abstract:The failure of deep neural networks to generalize to out-of-distribution data is a well-known problem and raises concerns about the deployment of trained networks in safety-critical domains such as healthcare, finance and autonomous vehicles. We study a particular kind of distribution shift $\unicode{x2013}$ shortcuts or spurious correlations in the training data. Shortcut learning is often only exposed when models are evaluated on real-world data that does not contain the same spurious correlations, posing a serious dilemma for AI practitioners to properly assess the effectiveness of a trained model for real-world applications. In this work, we propose to use the mutual information (MI) between the learned representation and the input as a metric to find where in training, the network latches onto shortcuts. Experiments demonstrate that MI can be used as a domain-agnostic metric for monitoring shortcut learning.
Domain-Agnostic Clustering with Self-Distillation
Nov 23, 2021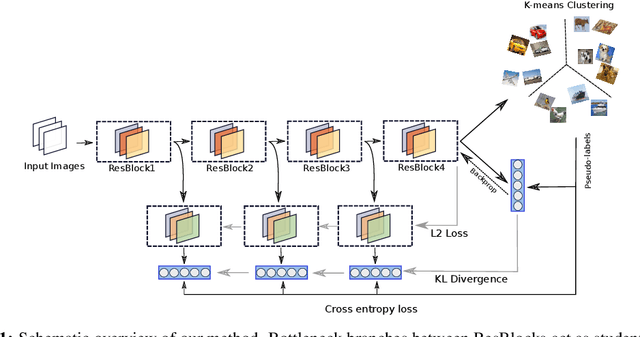
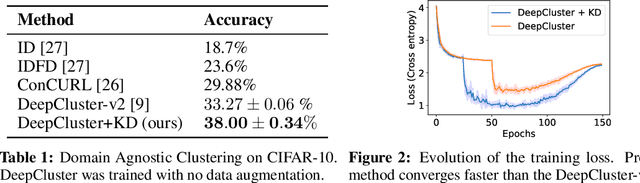
Abstract:Recent advancements in self-supervised learning have reduced the gap between supervised and unsupervised representation learning. However, most self-supervised and deep clustering techniques rely heavily on data augmentation, rendering them ineffective for many learning tasks where insufficient domain knowledge exists for performing augmentation. We propose a new self-distillation based algorithm for domain-agnostic clustering. Our method builds upon the existing deep clustering frameworks and requires no separate student model. The proposed method outperforms existing domain agnostic (augmentation-free) algorithms on CIFAR-10. We empirically demonstrate that knowledge distillation can improve unsupervised representation learning by extracting richer `dark knowledge' from the model than using predicted labels alone. Preliminary experiments also suggest that self-distillation improves the convergence of DeepCluster-v2.
Pay Attention with Focus: A Novel Learning Scheme for Classification of Whole Slide Images
Jun 11, 2021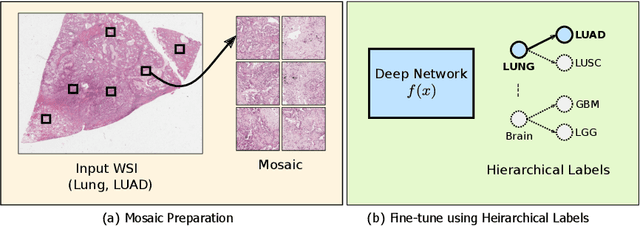
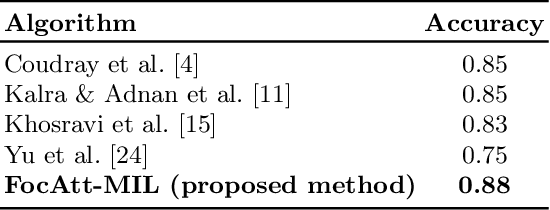
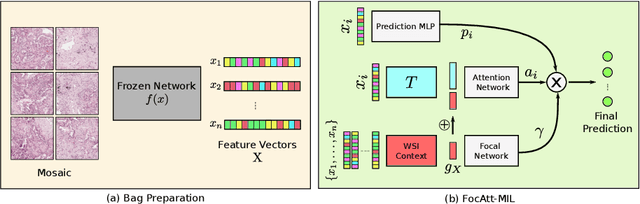
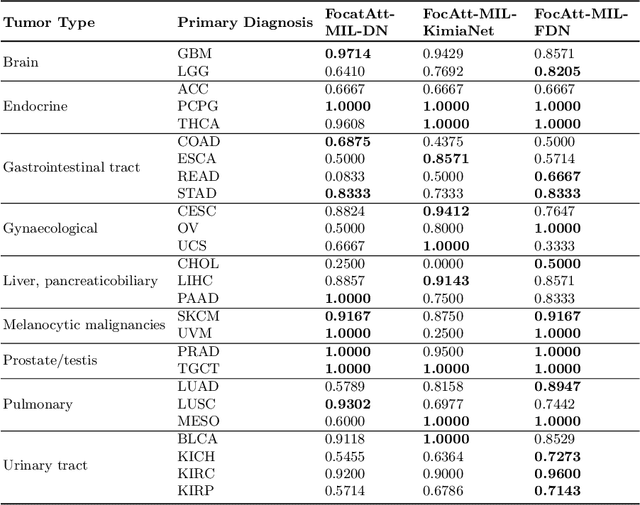
Abstract:Deep learning methods such as convolutional neural networks (CNNs) are difficult to directly utilize to analyze whole slide images (WSIs) due to the large image dimensions. We overcome this limitation by proposing a novel two-stage approach. First, we extract a set of representative patches (called mosaic) from a WSI. Each patch of a mosaic is encoded to a feature vector using a deep network. The feature extractor model is fine-tuned using hierarchical target labels of WSIs, i.e., anatomic site and primary diagnosis. In the second stage, a set of encoded patch-level features from a WSI is used to compute the primary diagnosis probability through the proposed Pay Attention with Focus scheme, an attention-weighted averaging of predicted probabilities for all patches of a mosaic modulated by a trainable focal factor. Experimental results show that the proposed model can be robust, and effective for the classification of WSIs.
Representation Learning of Histopathology Images using Graph Neural Networks
Apr 17, 2020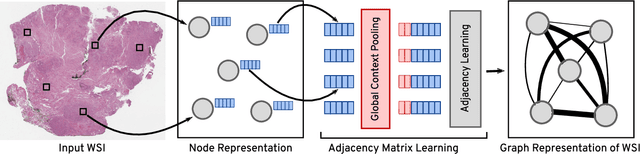
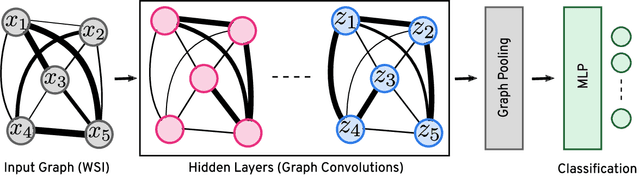
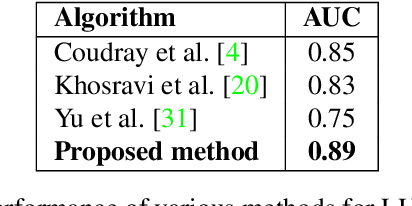
Abstract:Representation learning for Whole Slide Images (WSIs) is pivotal in developing image-based systems to achieve higher precision in diagnostic pathology. We propose a two-stage framework for WSI representation learning. We sample relevant patches using a color-based method and use graph neural networks to learn relations among sampled patches to aggregate the image information into a single vector representation. We introduce attention via graph pooling to automatically infer patches with higher relevance. We demonstrate the performance of our approach for discriminating two sub-types of lung cancers, Lung Adenocarcinoma (LUAD) & Lung Squamous Cell Carcinoma (LUSC). We collected 1,026 lung cancer WSIs with the 40$\times$ magnification from The Cancer Genome Atlas (TCGA) dataset, the largest public repository of histopathology images and achieved state-of-the-art accuracy of 88.8% and AUC of 0.89 on lung cancer sub-type classification by extracting features from a pre-trained DenseNet
Learning Permutation Invariant Representations using Memory Networks
Nov 18, 2019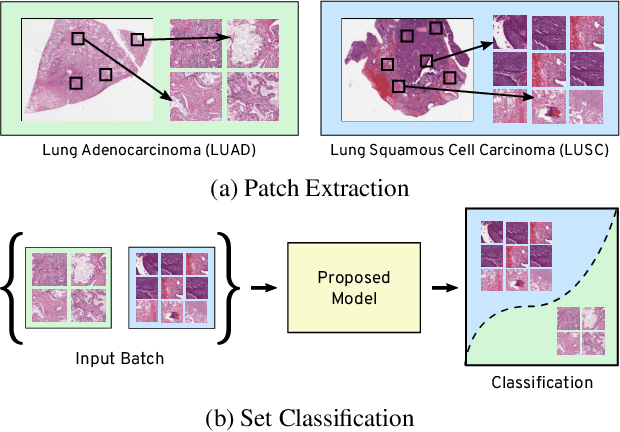
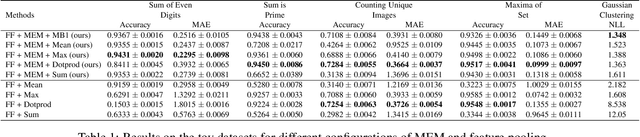
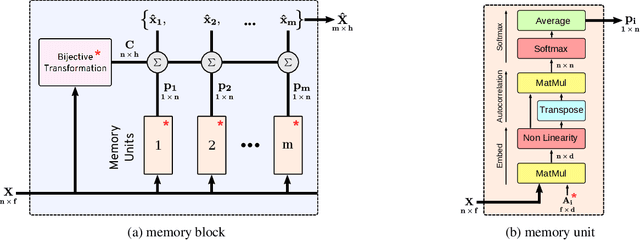

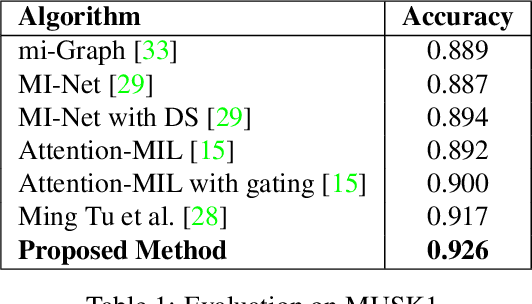
Abstract:Many real world tasks such as 3D object detection and high-resolution image classification involve learning from a set of instances. In these cases, only a group of instances, a set, collectively contains meaningful information and therefore only the sets have labels, and not individual data instances. In this work, we present a permutation invariant neural network called a \textbf{Memory-based Exchangeable Model (MEM)} for learning set functions. The model consists of memory units that embed an input sequence to high-level features (memories) enabling the model to learn inter-dependencies among instances of the set in the form of attention vectors. To demonstrate its learning ability, we evaluated our model on test datasets created using MNIST, point cloud classification, and population estimation. We also tested the model for classifying histopathology whole slide images to discriminate between two subtypes of Lung cancer---Lung Adenocarcinoma, and Lung Squamous Cell Carcinoma. We systematically extracted patches from lung cancer images from The Cancer Genome Atlas~(TCGA) dataset, the largest public repository of histopathology images. The proposed method achieved a competitive classification accuracy of 84.84\%. The results on other datasets are promising and demonstrate the efficacy of our model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge