Hamid Tizhoosh
Pay Attention with Focus: A Novel Learning Scheme for Classification of Whole Slide Images
Jun 11, 2021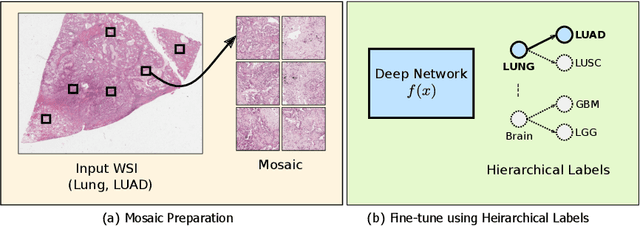
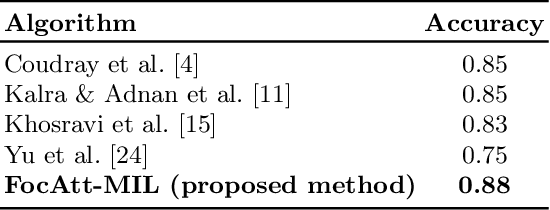
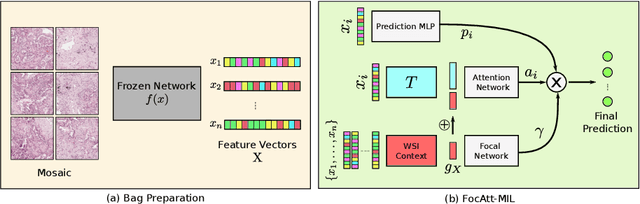
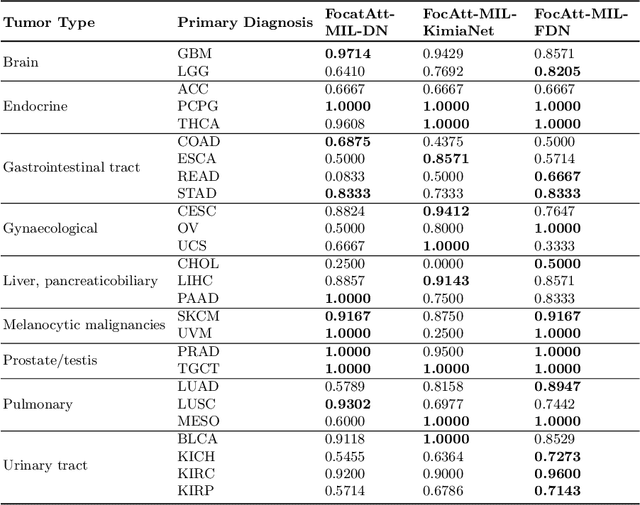
Abstract:Deep learning methods such as convolutional neural networks (CNNs) are difficult to directly utilize to analyze whole slide images (WSIs) due to the large image dimensions. We overcome this limitation by proposing a novel two-stage approach. First, we extract a set of representative patches (called mosaic) from a WSI. Each patch of a mosaic is encoded to a feature vector using a deep network. The feature extractor model is fine-tuned using hierarchical target labels of WSIs, i.e., anatomic site and primary diagnosis. In the second stage, a set of encoded patch-level features from a WSI is used to compute the primary diagnosis probability through the proposed Pay Attention with Focus scheme, an attention-weighted averaging of predicted probabilities for all patches of a mosaic modulated by a trainable focal factor. Experimental results show that the proposed model can be robust, and effective for the classification of WSIs.
Searching for Pneumothorax in Half a Million Chest X-Ray Images
Jul 30, 2020



Abstract:Pneumothorax, a collapsed or dropped lung, is a fatal condition typically detected on a chest X-ray by an experienced radiologist. Due to shortage of such experts, automated detection systems based on deep neural networks have been developed. Nevertheless, applying such systems in practice remains a challenge. These systems, mostly compute a single probability as output, may not be enough for diagnosis. On the contrary, content-based medical image retrieval (CBIR) systems, such as image search, can assist clinicians for diagnostic purposes by enabling them to compare the case they are examining with previous (already diagnosed) cases. However, there is a lack of study on such attempt. In this study, we explored the use of image search to classify pneumothorax among chest X-ray images. All chest X-ray images were first tagged with deep pretrained features, which were obtained from existing deep learning models. Given a query chest X-ray image, the majority voting of the top K retrieved images was then used as a classifier, in which similar cases in the archive of past cases are provided besides the probability output. In our experiments, 551,383 chest X-ray images were obtained from three large recently released public datasets. Using 10-fold cross-validation, it is shown that image search on deep pretrained features achieved promising results compared to those obtained by traditional classifiers trained on the same features. To the best of knowledge, it is the first study to demonstrate that deep pretrained features can be used for CBIR of pneumothorax in half a million chest X-ray images.
Learning Permutation Invariant Representations using Memory Networks
Nov 18, 2019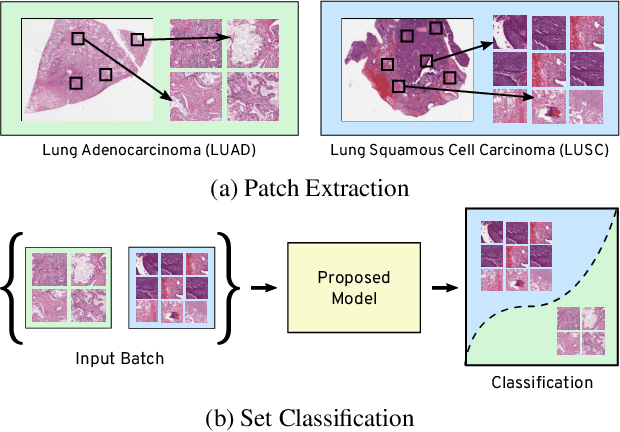
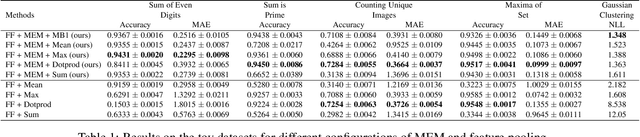
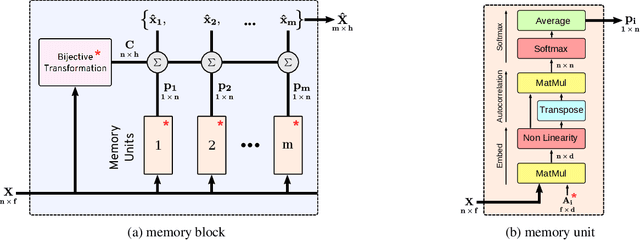

Abstract:Many real world tasks such as 3D object detection and high-resolution image classification involve learning from a set of instances. In these cases, only a group of instances, a set, collectively contains meaningful information and therefore only the sets have labels, and not individual data instances. In this work, we present a permutation invariant neural network called a \textbf{Memory-based Exchangeable Model (MEM)} for learning set functions. The model consists of memory units that embed an input sequence to high-level features (memories) enabling the model to learn inter-dependencies among instances of the set in the form of attention vectors. To demonstrate its learning ability, we evaluated our model on test datasets created using MNIST, point cloud classification, and population estimation. We also tested the model for classifying histopathology whole slide images to discriminate between two subtypes of Lung cancer---Lung Adenocarcinoma, and Lung Squamous Cell Carcinoma. We systematically extracted patches from lung cancer images from The Cancer Genome Atlas~(TCGA) dataset, the largest public repository of histopathology images. The proposed method achieved a competitive classification accuracy of 84.84\%. The results on other datasets are promising and demonstrate the efficacy of our model.
A Compact Representation of Histopathology Images using Digital Stain Separation & Frequency-Based Encoded Local Projections
May 28, 2019



Abstract:In recent years, histopathology images have been increasingly used as a diagnostic tool in the medical field. The process of accurately diagnosing a biopsy sample requires significant expertise in the field, and as such can be time-consuming and is prone to uncertainty and error. With the advent of digital pathology, using image recognition systems to highlight problem areas or locate similar images can aid pathologists in making quick and accurate diagnoses. In this paper, we specifically consider the encoded local projections (ELP) algorithm, which has previously shown some success as a tool for classification and recognition of histopathology images. We build on the success of the ELP algorithm as a means for image classification and recognition by proposing a modified algorithm which captures the local frequency information of the image. The proposed algorithm estimates local frequencies by quantifying the changes in multiple projections in local windows of greyscale images. By doing so we remove the need to store the full projections, thus significantly reducing the histogram size, and decreasing computation time for image retrieval and classification tasks. Furthermore, we investigate the effectiveness of applying our method to histopathology images which have been digitally separated into their hematoxylin and eosin stain components. The proposed algorithm is tested on the publicly available invasive ductal carcinoma (IDC) data set. The histograms are used to train an SVM to classify the data. The experiments showed that the proposed method outperforms the original ELP algorithm in image retrieval tasks. On classification tasks, the results are found to be comparable to state-of-the-art deep learning methods and better than many handcrafted features from the literature.
Deep Barcodes for Fast Retrieval of Histopathology Scans
Apr 30, 2018
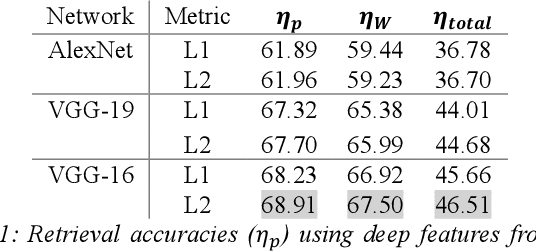
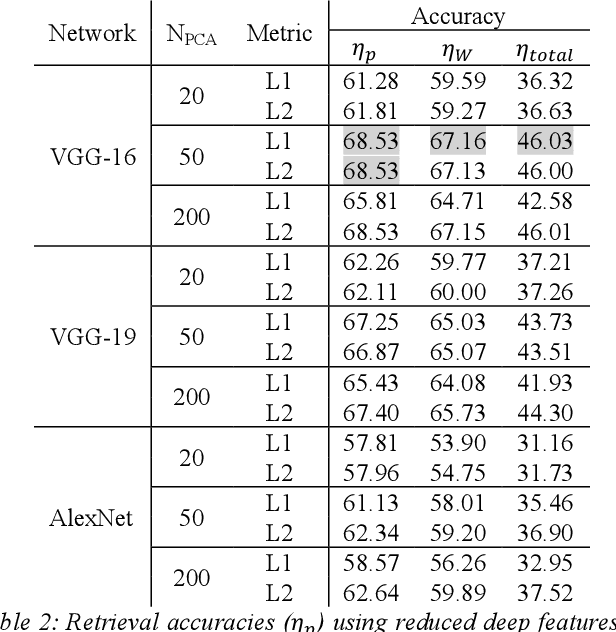
Abstract:We investigate the concept of deep barcodes and propose two methods to generate them in order to expedite the process of classification and retrieval of histopathology images. Since binary search is computationally less expensive, in terms of both speed and storage, deep barcodes could be useful when dealing with big data retrieval. Our experiments use the dataset Kimia Path24 to test three pre-trained networks for image retrieval. The dataset consists of 27,055 training images in 24 different classes with large variability, and 1,325 test images for testing. Apart from the high-speed and efficiency, results show a surprising retrieval accuracy of 71.62% for deep barcodes, as compared to 68.91% for deep features and 68.53% for compressed deep features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge