Meghana Dinesh Kumar
Deep Barcodes for Fast Retrieval of Histopathology Scans
Apr 30, 2018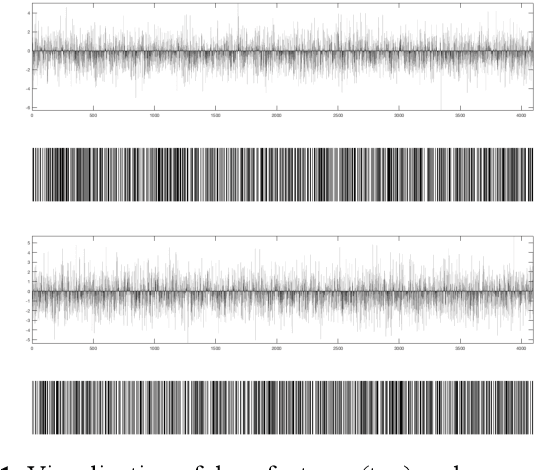
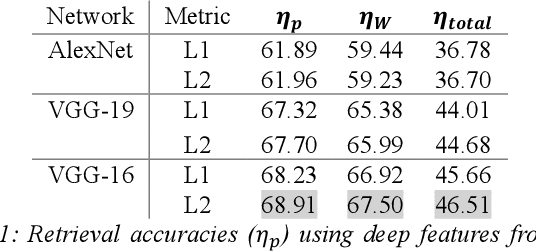
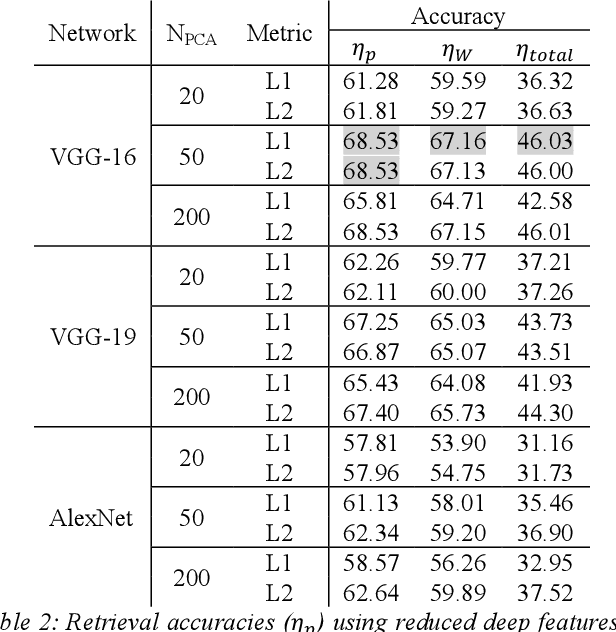
Abstract:We investigate the concept of deep barcodes and propose two methods to generate them in order to expedite the process of classification and retrieval of histopathology images. Since binary search is computationally less expensive, in terms of both speed and storage, deep barcodes could be useful when dealing with big data retrieval. Our experiments use the dataset Kimia Path24 to test three pre-trained networks for image retrieval. The dataset consists of 27,055 training images in 24 different classes with large variability, and 1,325 test images for testing. Apart from the high-speed and efficiency, results show a surprising retrieval accuracy of 71.62% for deep barcodes, as compared to 68.91% for deep features and 68.53% for compressed deep features.
A Comparative Study of CNN, BoVW and LBP for Classification of Histopathological Images
Sep 27, 2017



Abstract:Despite the progress made in the field of medical imaging, it remains a large area of open research, especially due to the variety of imaging modalities and disease-specific characteristics. This paper is a comparative study describing the potential of using local binary patterns (LBP), deep features and the bag-of-visual words (BoVW) scheme for the classification of histopathological images. We introduce a new dataset, \emph{KIMIA Path960}, that contains 960 histopathology images belonging to 20 different classes (different tissue types). We make this dataset publicly available. The small size of the dataset and its inter- and intra-class variability makes it ideal for initial investigations when comparing image descriptors for search and classification in complex medical imaging cases like histopathology. We investigate deep features, LBP histograms and BoVW to classify the images via leave-one-out validation. The accuracy of image classification obtained using LBP was 90.62\% while the highest accuracy using deep features reached 94.72\%. The dictionary approach (BoVW) achieved 96.50\%. Deep solutions may be able to deliver higher accuracies but they need extensive training with a large number of (balanced) image datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge