Taher Dehkharghanian
Evolutionary Computation in Action: Hyperdimensional Deep Embedding Spaces of Gigapixel Pathology Images
Mar 02, 2023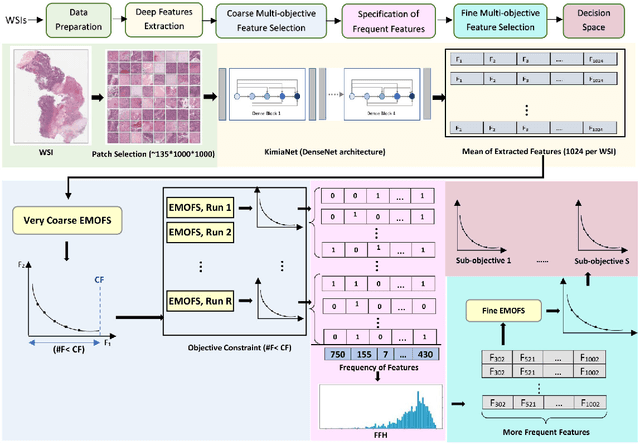



Abstract:One of the main obstacles of adopting digital pathology is the challenge of efficient processing of hyperdimensional digitized biopsy samples, called whole slide images (WSIs). Exploiting deep learning and introducing compact WSI representations are urgently needed to accelerate image analysis and facilitate the visualization and interpretability of pathology results in a postpandemic world. In this paper, we introduce a new evolutionary approach for WSI representation based on large-scale multi-objective optimization (LSMOP) of deep embeddings. We start with patch-based sampling to feed KimiaNet , a histopathology-specialized deep network, and to extract a multitude of feature vectors. Coarse multi-objective feature selection uses the reduced search space strategy guided by the classification accuracy and the number of features. In the second stage, the frequent features histogram (FFH), a novel WSI representation, is constructed by multiple runs of coarse LSMOP. Fine evolutionary feature selection is then applied to find a compact (short-length) feature vector based on the FFH and contributes to a more robust deep-learning approach to digital pathology supported by the stochastic power of evolutionary algorithms. We validate the proposed schemes using The Cancer Genome Atlas (TCGA) images in terms of WSI representation, classification accuracy, and feature quality. Furthermore, a novel decision space for multicriteria decision making in the LSMOP field is introduced. Finally, a patch-level visualization approach is proposed to increase the interpretability of deep features. The proposed evolutionary algorithm finds a very compact feature vector to represent a WSI (almost 14,000 times smaller than the original feature vectors) with 8% higher accuracy compared to the codes provided by the state-of-the-art methods.
Histogram of Cell Types: Deep Learning for Automated Bone Marrow Cytology
Jul 08, 2021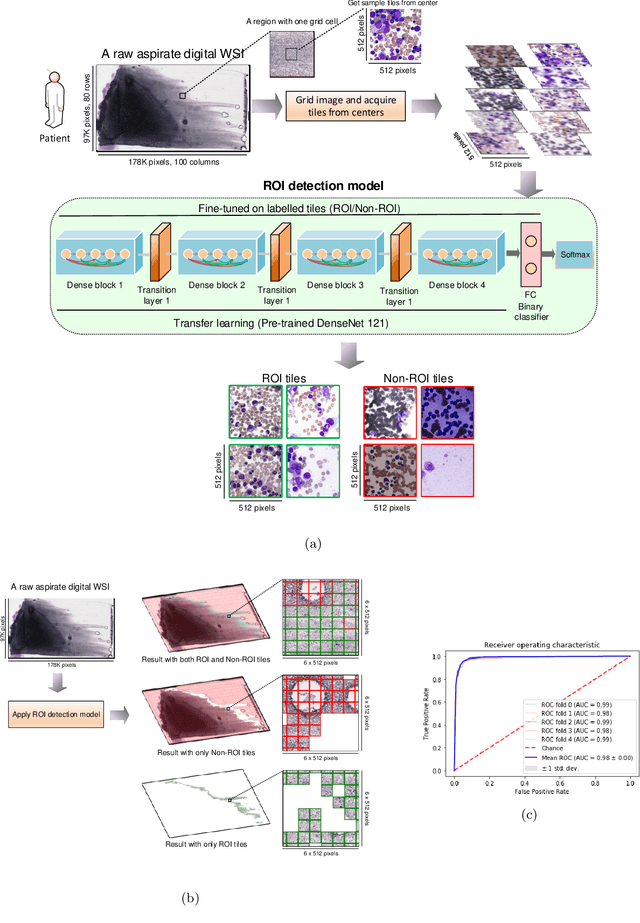
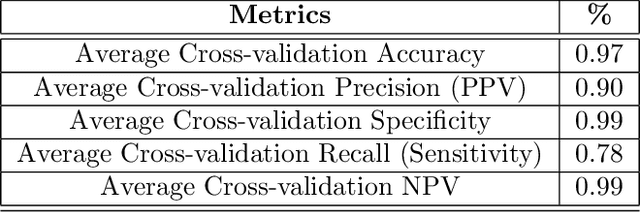
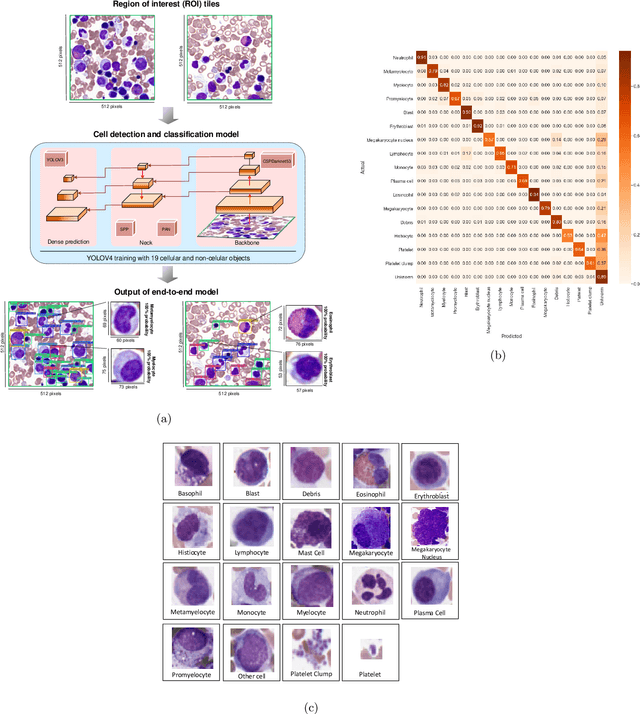
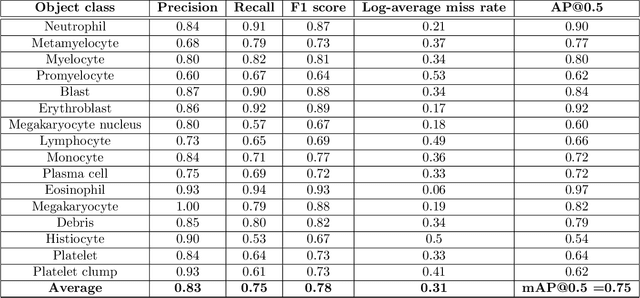
Abstract:Bone marrow cytology is required to make a hematological diagnosis, influencing critical clinical decision points in hematology. However, bone marrow cytology is tedious, limited to experienced reference centers and associated with high inter-observer variability. This may lead to a delayed or incorrect diagnosis, leaving an unmet need for innovative supporting technologies. We have developed the first ever end-to-end deep learning-based technology for automated bone marrow cytology. Starting with a bone marrow aspirate digital whole slide image, our technology rapidly and automatically detects suitable regions for cytology, and subsequently identifies and classifies all bone marrow cells in each region. This collective cytomorphological information is captured in a novel representation called Histogram of Cell Types (HCT) quantifying bone marrow cell class probability distribution and acting as a cytological "patient fingerprint". The approach achieves high accuracy in region detection (0.97 accuracy and 0.99 ROC AUC), and cell detection and cell classification (0.75 mAP, 0.78 F1-score, Log-average miss rate of 0.31). HCT has potential to revolutionize hematopathology diagnostic workflows, leading to more cost-effective, accurate diagnosis and opening the door to precision medicine.
Pay Attention with Focus: A Novel Learning Scheme for Classification of Whole Slide Images
Jun 11, 2021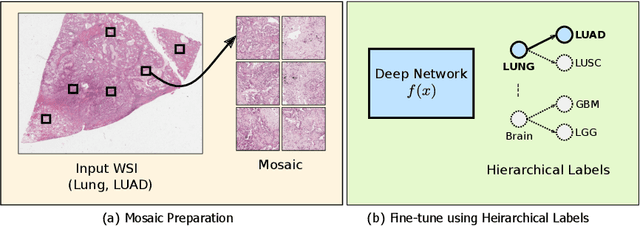
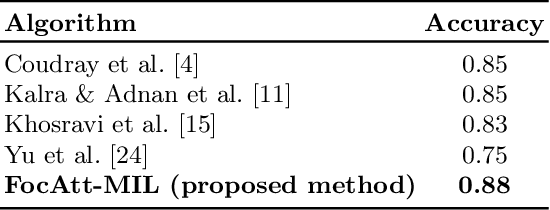
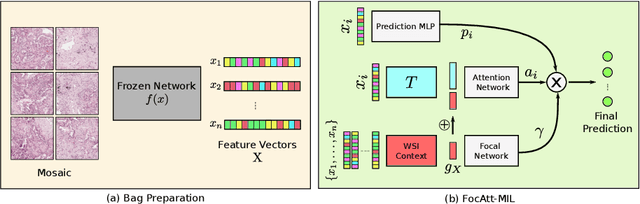
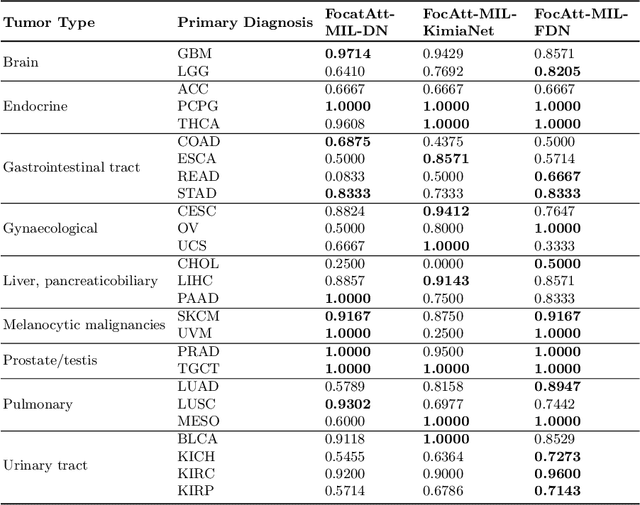
Abstract:Deep learning methods such as convolutional neural networks (CNNs) are difficult to directly utilize to analyze whole slide images (WSIs) due to the large image dimensions. We overcome this limitation by proposing a novel two-stage approach. First, we extract a set of representative patches (called mosaic) from a WSI. Each patch of a mosaic is encoded to a feature vector using a deep network. The feature extractor model is fine-tuned using hierarchical target labels of WSIs, i.e., anatomic site and primary diagnosis. In the second stage, a set of encoded patch-level features from a WSI is used to compute the primary diagnosis probability through the proposed Pay Attention with Focus scheme, an attention-weighted averaging of predicted probabilities for all patches of a mosaic modulated by a trainable focal factor. Experimental results show that the proposed model can be robust, and effective for the classification of WSIs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge