Hamid R. Tizhoosh
A Preliminary Investigation into Search and Matching for Tumour Discrimination in WHO Breast Taxonomy Using Deep Networks
Aug 22, 2023Abstract:Breast cancer is one of the most common cancers affecting women worldwide. They include a group of malignant neoplasms with a variety of biological, clinical, and histopathological characteristics. There are more than 35 different histological forms of breast lesions that can be classified and diagnosed histologically according to cell morphology, growth, and architecture patterns. Recently, deep learning, in the field of artificial intelligence, has drawn a lot of attention for the computerized representation of medical images. Searchable digital atlases can provide pathologists with patch matching tools allowing them to search among evidently diagnosed and treated archival cases, a technology that may be regarded as computational second opinion. In this study, we indexed and analyzed the WHO breast taxonomy (Classification of Tumours 5th Ed.) spanning 35 tumour types. We visualized all tumour types using deep features extracted from a state-of-the-art deep learning model, pre-trained on millions of diagnostic histopathology images from the TCGA repository. Furthermore, we test the concept of a digital "atlas" as a reference for search and matching with rare test cases. The patch similarity search within the WHO breast taxonomy data reached over 88% accuracy when validating through "majority vote" and more than 91% accuracy when validating using top-n tumour types. These results show for the first time that complex relationships among common and rare breast lesions can be investigated using an indexed digital archive.
ProxyFL: Decentralized Federated Learning through Proxy Model Sharing
Nov 22, 2021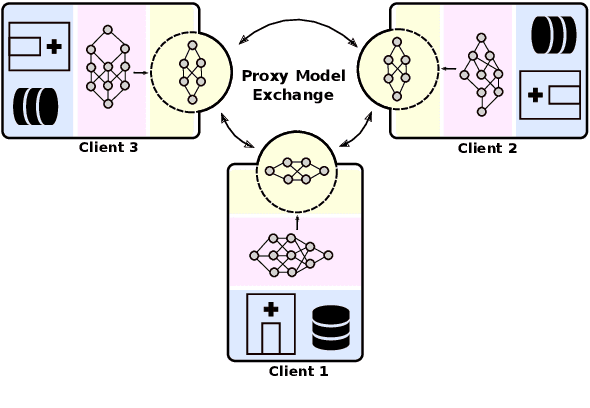
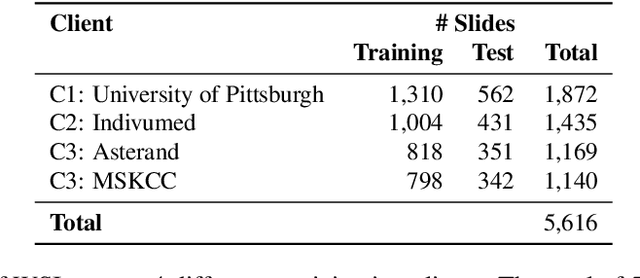
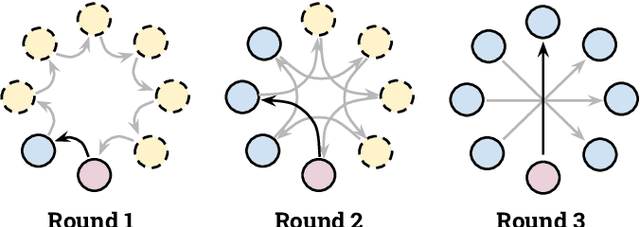
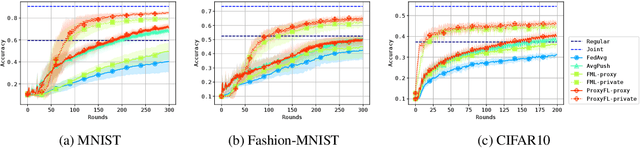
Abstract:Institutions in highly regulated domains such as finance and healthcare often have restrictive rules around data sharing. Federated learning is a distributed learning framework that enables multi-institutional collaborations on decentralized data with improved protection for each collaborator's data privacy. In this paper, we propose a communication-efficient scheme for decentralized federated learning called ProxyFL, or proxy-based federated learning. Each participant in ProxyFL maintains two models, a private model, and a publicly shared proxy model designed to protect the participant's privacy. Proxy models allow efficient information exchange among participants using the PushSum method without the need of a centralized server. The proposed method eliminates a significant limitation of canonical federated learning by allowing model heterogeneity; each participant can have a private model with any architecture. Furthermore, our protocol for communication by proxy leads to stronger privacy guarantees using differential privacy analysis. Experiments on popular image datasets, and a pan-cancer diagnostic problem using over 30,000 high-quality gigapixel histology whole slide images, show that ProxyFL can outperform existing alternatives with much less communication overhead and stronger privacy.
Histogram of Cell Types: Deep Learning for Automated Bone Marrow Cytology
Jul 08, 2021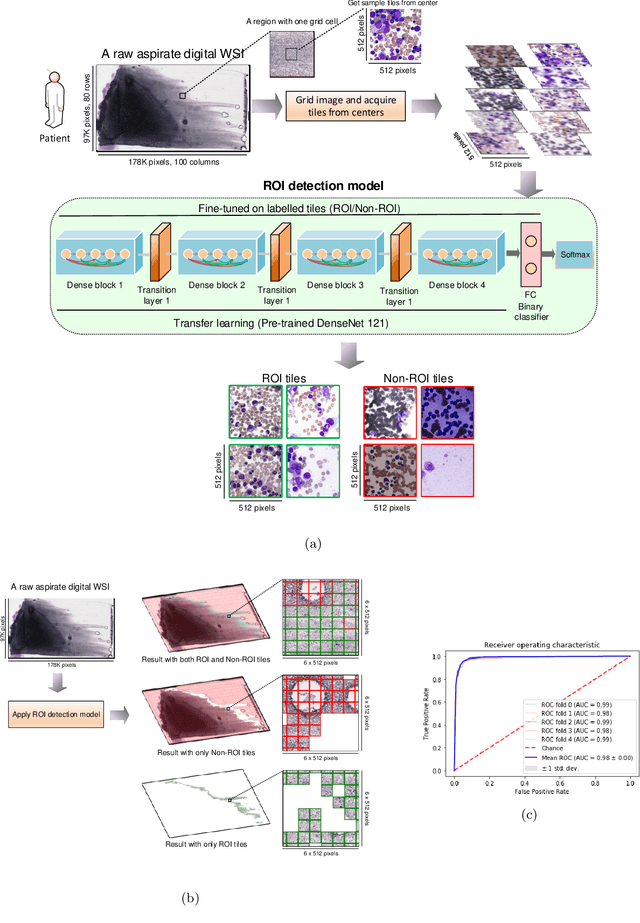
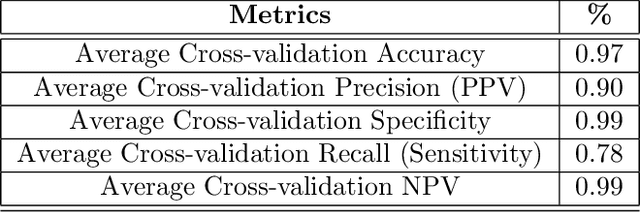
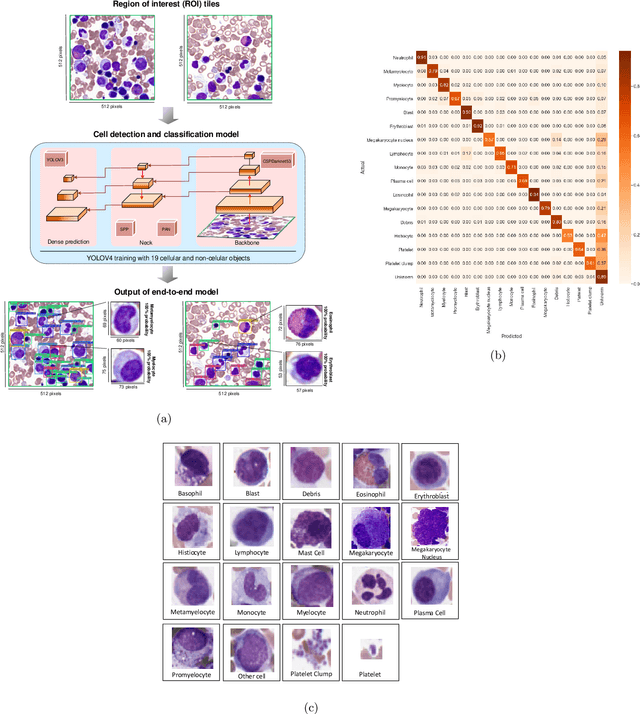
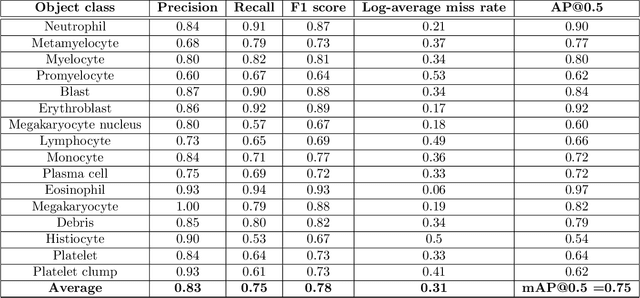
Abstract:Bone marrow cytology is required to make a hematological diagnosis, influencing critical clinical decision points in hematology. However, bone marrow cytology is tedious, limited to experienced reference centers and associated with high inter-observer variability. This may lead to a delayed or incorrect diagnosis, leaving an unmet need for innovative supporting technologies. We have developed the first ever end-to-end deep learning-based technology for automated bone marrow cytology. Starting with a bone marrow aspirate digital whole slide image, our technology rapidly and automatically detects suitable regions for cytology, and subsequently identifies and classifies all bone marrow cells in each region. This collective cytomorphological information is captured in a novel representation called Histogram of Cell Types (HCT) quantifying bone marrow cell class probability distribution and acting as a cytological "patient fingerprint". The approach achieves high accuracy in region detection (0.97 accuracy and 0.99 ROC AUC), and cell detection and cell classification (0.75 mAP, 0.78 F1-score, Log-average miss rate of 0.31). HCT has potential to revolutionize hematopathology diagnostic workflows, leading to more cost-effective, accurate diagnosis and opening the door to precision medicine.
Searching for Pneumothorax in X-Ray Images Using Autoencoded Deep Features
Feb 11, 2021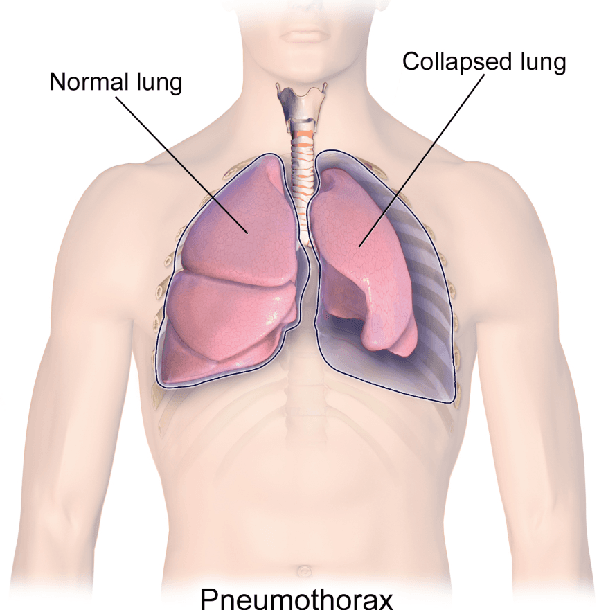

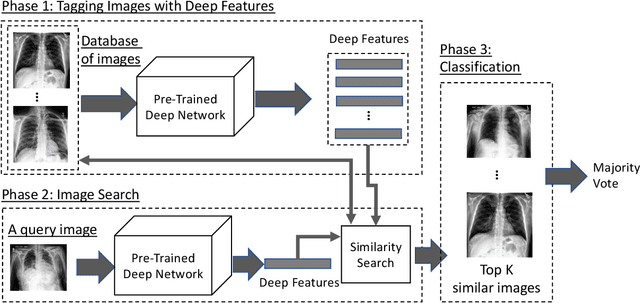
Abstract:Fast diagnosis and treatment of pneumothorax, a collapsed or dropped lung, is crucial to avoid fatalities. Pneumothorax is typically detected on a chest X-ray image through visual inspection by experienced radiologists. However, the detection rate is quite low. Therefore, there is a strong need for automated detection systems to assist radiologists. Despite the high accuracy levels generally reported for deep learning classifiers in many applications, they may not be useful in clinical practice due to the lack of large number of high-quality labelled images as well as a lack of interpretation possibility. Alternatively, searching in the archive of past cases to find matching images may serve as a 'virtual second opinion' through accessing the metadata of matched evidently diagnosed cases. To use image search as a triaging/diagnosis tool, all chest X-ray images must first be tagged with identifiers, i.e., deep features. Then, given a query chest X-ray image, the majority vote among the top k retrieved images can provide a more explainable output. While image search can be clinically more viable, its detection performance needs to be investigated at a scale closer to real-world practice. We combined 3 public datasets to assemble a repository with more than 550,000 chest X-ray images. We developed the Autoencoding Thorax Net (short AutoThorax-Net) for image search in chest radiographs compressing three inputs: the left chest side, the flipped right side, and the entire chest image. Experimental results show that image search based on AutoThorax-Net features can achieve high identification rates providing a path towards real-world deployment. We achieved 92% AUC accuracy for a semi-automated search in 194,608 images (pneumothorax and normal) and 82% AUC accuracy for fully automated search in 551,383 images (normal, pneumothorax and many other chest diseases).
Forming Local Intersections of Projections for Classifying and Searching Histopathology Images
Aug 08, 2020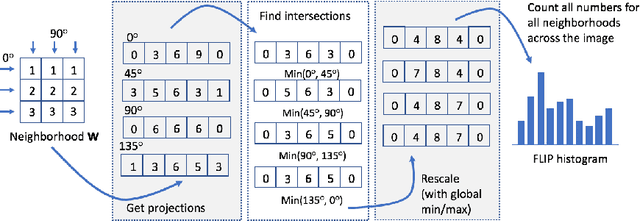
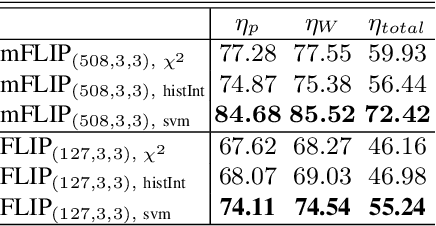
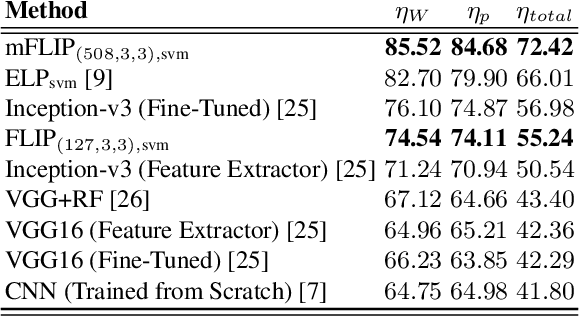

Abstract:In this paper, we propose a novel image descriptor called Forming Local Intersections of Projections (FLIP) and its multi-resolution version (mFLIP) for representing histopathology images. The descriptor is based on the Radon transform wherein we apply parallel projections in small local neighborhoods of gray-level images. Using equidistant projection directions in each window, we extract unique and invariant characteristics of the neighborhood by taking the intersection of adjacent projections. Thereafter, we construct a histogram for each image, which we call the FLIP histogram. Various resolutions provide different FLIP histograms which are then concatenated to form the mFLIP descriptor. Our experiments included training common networks from scratch and fine-tuning pre-trained networks to benchmark our proposed descriptor. Experiments are conducted on the publicly available dataset KIMIA Path24 and KIMIA Path960. For both of these datasets, FLIP and mFLIP descriptors show promising results in all experiments.Using KIMIA Path24 data, FLIP outperformed non-fine-tuned Inception-v3 and fine-tuned VGG16 and mFLIP outperformed fine-tuned Inception-v3 in feature extracting.
Representation Learning of Histopathology Images using Graph Neural Networks
Apr 17, 2020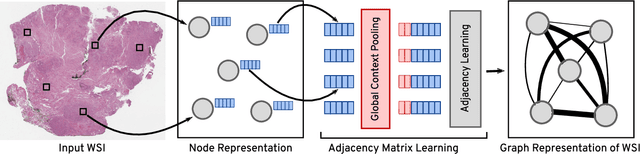
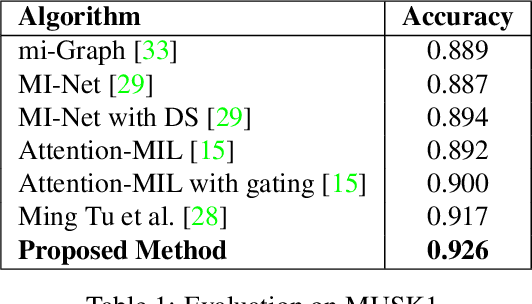
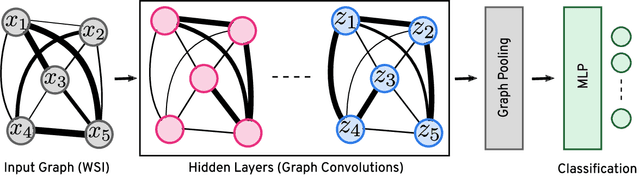
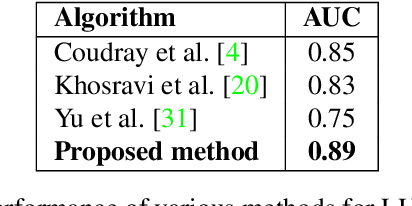
Abstract:Representation learning for Whole Slide Images (WSIs) is pivotal in developing image-based systems to achieve higher precision in diagnostic pathology. We propose a two-stage framework for WSI representation learning. We sample relevant patches using a color-based method and use graph neural networks to learn relations among sampled patches to aggregate the image information into a single vector representation. We introduce attention via graph pooling to automatically infer patches with higher relevance. We demonstrate the performance of our approach for discriminating two sub-types of lung cancers, Lung Adenocarcinoma (LUAD) & Lung Squamous Cell Carcinoma (LUSC). We collected 1,026 lung cancer WSIs with the 40$\times$ magnification from The Cancer Genome Atlas (TCGA) dataset, the largest public repository of histopathology images and achieved state-of-the-art accuracy of 88.8% and AUC of 0.89 on lung cancer sub-type classification by extracting features from a pre-trained DenseNet
Automatic Classification of Pathology Reports using TF-IDF Features
Mar 05, 2019



Abstract:A Pathology report is arguably one of the most important documents in medicine containing interpretive information about the visual findings from the patient's biopsy sample. Each pathology report has a retention period of up to 20 years after the treatment of a patient. Cancer registries process and encode high volumes of free-text pathology reports for surveillance of cancer and tumor diseases all across the world. In spite of their extremely valuable information they hold, pathology reports are not used in any systematic way to facilitate computational pathology. Therefore, in this study, we investigate automated machine-learning techniques to identify/predict the primary diagnosis (based on ICD-O code) from pathology reports. We performed experiments by extracting the TF-IDF features from the reports and classifying them using three different methods---SVM, XGBoost, and Logistic Regression. We constructed a new dataset with 1,949 pathology reports arranged into 37 ICD-O categories, collected from four different primary sites, namely lung, kidney, thymus, and testis. The reports were manually transcribed into text format after collecting them as PDF files from NCI Genomic Data Commons public dataset. We subsequently pre-processed the reports by removing irrelevant textual artifacts produced by OCR software. The highest classification accuracy we achieved was 92\% using XGBoost classifier on TF-IDF feature vectors, the linear SVM scored 87\% accuracy. Furthermore, the study shows that TF-IDF vectors are suitable for highlighting the important keywords within a report which can be helpful for the cancer research and diagnostic workflow. The results are encouraging in demonstrating the potential of machine learning methods for classification and encoding of pathology reports.
A Dataset and Preliminary Results for Umpire Pose Detection Using SVM Classification of Deep Features
Sep 11, 2018

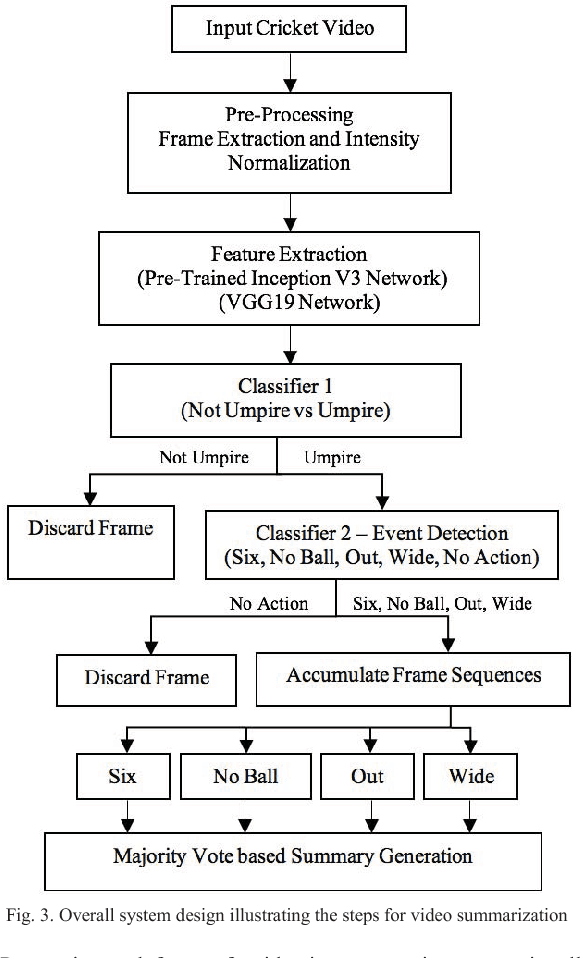

Abstract:In recent years, there has been increased interest in video summarization and automatic sports highlights generation. In this work, we introduce a new dataset, called SNOW, for umpire pose detection in the game of cricket. The proposed dataset is evaluated as a preliminary aid for developing systems to automatically generate cricket highlights. In cricket, the umpire has the authority to make important decisions about events on the field. The umpire signals important events using unique hand signals and gestures. We identify four such events for classification namely SIX, NO BALL, OUT and WIDE based on detecting the pose of the umpire from the frames of a cricket video. Pre-trained convolutional neural networks such as Inception V3 and VGG19 net-works are selected as primary candidates for feature extraction. The results are obtained using a linear SVM classifier. The highest classification performance was achieved for the SVM trained on features extracted from the VGG19 network. The preliminary results suggest that the proposed system is an effective solution for the application of cricket highlights generation.
Opposition based Ensemble Micro Differential Evolution
Sep 21, 2017



Abstract:Differential evolution (DE) algorithm with a small population size is called Micro-DE (MDE). A small population size decreases the computational complexity but also reduces the exploration ability of DE by limiting the population diversity. In this paper, we propose the idea of combining ensemble mutation scheme selection and opposition-based learning concepts to enhance the diversity of population in MDE at mutation and selection stages. The proposed algorithm enhances the diversity of population by generating a random mutation scale factor per individual and per dimension, randomly assigning a mutation scheme to each individual in each generation, and diversifying individuals selection using opposition-based learning. This approach is easy to implement and does not require the setting of mutation scheme selection and mutation scale factor. Experimental results are conducted for a variety of objective functions with low and high dimensionality on the CEC Black- Box Optimization Benchmarking 2015 (CEC-BBOB 2015). The results show superior performance of the proposed algorithm compared to the other micro-DE algorithms.
Tumour Ellipsification in Ultrasound Images for Treatment Prediction in Breast Cancer
Jan 13, 2017
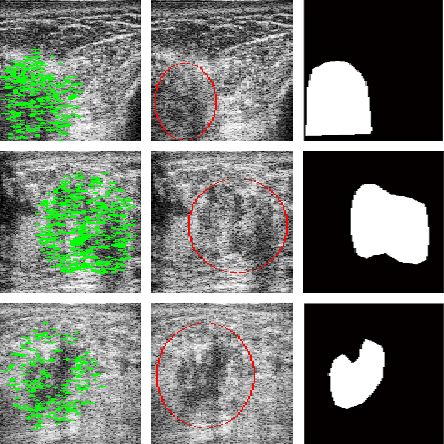
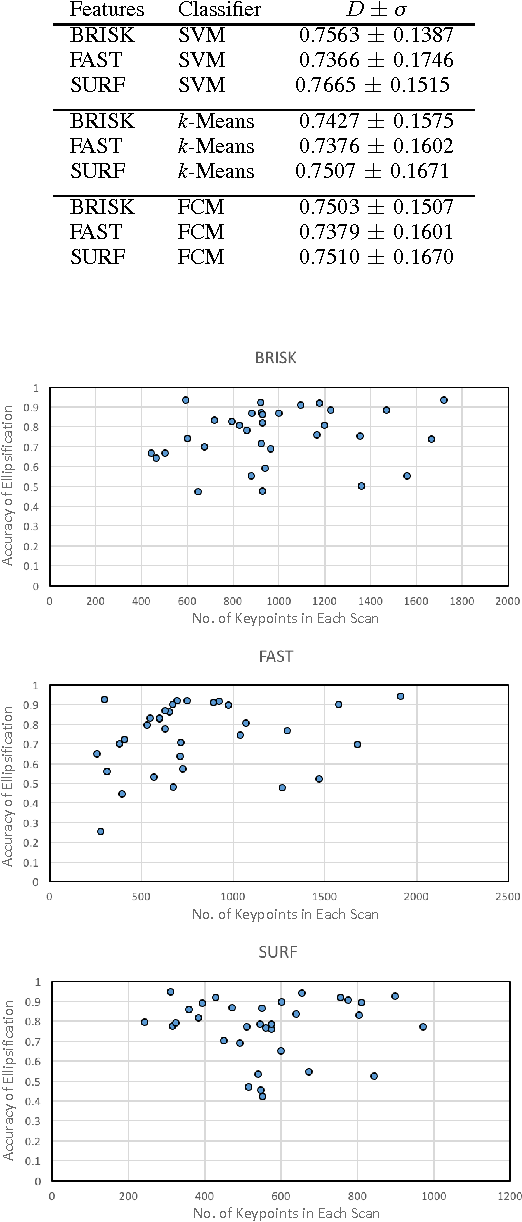
Abstract:Recent advances in using quantitative ultrasound (QUS) methods have provided a promising framework to non-invasively and inexpensively monitor or predict the effectiveness of therapeutic cancer responses. One of the earliest steps in using QUS methods is contouring a region of interest (ROI) inside the tumour in ultrasound B-mode images. While manual segmentation is a very time-consuming and tedious task for human experts, auto-contouring is also an extremely difficult task for computers due to the poor quality of ultrasound B-mode images. However, for the purpose of cancer response prediction, a rough boundary of the tumour as an ROI is only needed. In this research, a semi-automated tumour localization approach is proposed for ROI estimation in ultrasound B-mode images acquired from patients with locally advanced breast cancer (LABC). The proposed approach comprised several modules, including 1) feature extraction using keypoint descriptors, 2) augmenting the feature descriptors with the distance of the keypoints to the user-input pixel as the centre of the tumour, 3) supervised learning using a support vector machine (SVM) to classify keypoints as "tumour" or "non-tumour", and 4) computation of an ellipse as an outline of the ROI representing the tumour. Experiments with 33 B-mode images from 10 LABC patients yielded promising results with an accuracy of 76.7% based on the Dice coefficient performance measure. The results demonstrated that the proposed method can potentially be used as the first stage in a computer-assisted cancer response prediction system for semi-automated contouring of breast tumours.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge