Danial Maleki
LILE: Look In-Depth before Looking Elsewhere -- A Dual Attention Network using Transformers for Cross-Modal Information Retrieval in Histopathology Archives
Mar 04, 2022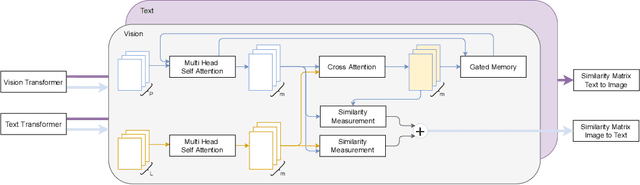
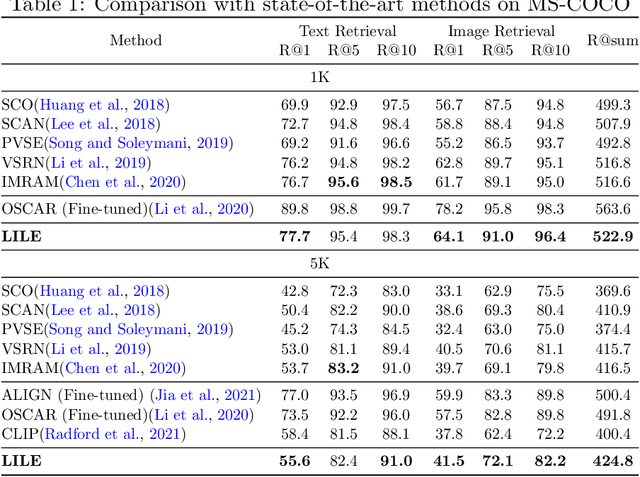
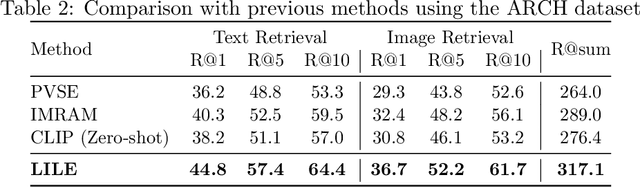
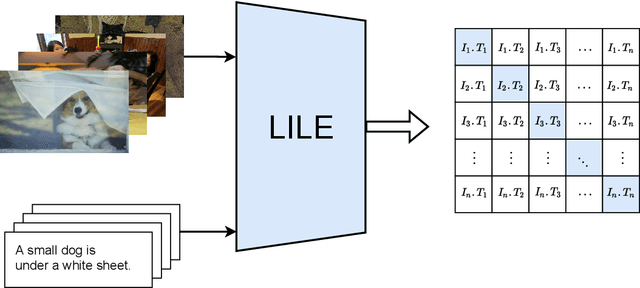
Abstract:The volume of available data has grown dramatically in recent years in many applications. Furthermore, the age of networks that used multiple modalities separately has practically ended. Therefore, enabling bidirectional cross-modality data retrieval capable of processing has become a requirement for many domains and disciplines of research. This is especially true in the medical field, as data comes in a multitude of types, including various types of images and reports as well as molecular data. Most contemporary works apply cross attention to highlight the essential elements of an image or text in relation to the other modalities and try to match them together. However, regardless of their importance in their own modality, these approaches usually consider features of each modality equally. In this study, self-attention as an additional loss term will be proposed to enrich the internal representation provided into the cross attention module. This work suggests a novel architecture with a new loss term to help represent images and texts in the joint latent space. Experiment results on two benchmark datasets, i.e. MS-COCO and ARCH, show the effectiveness of the proposed method.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021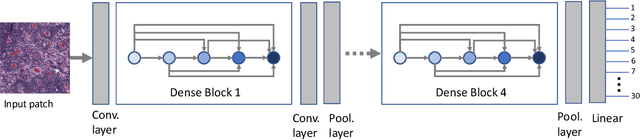
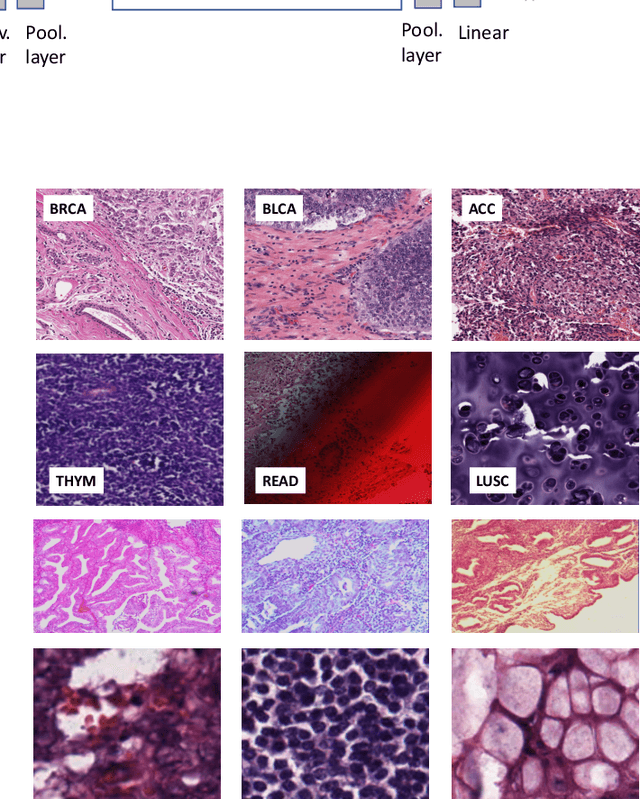
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Ink Marker Segmentation in Histopathology Images Using Deep Learning
Oct 29, 2020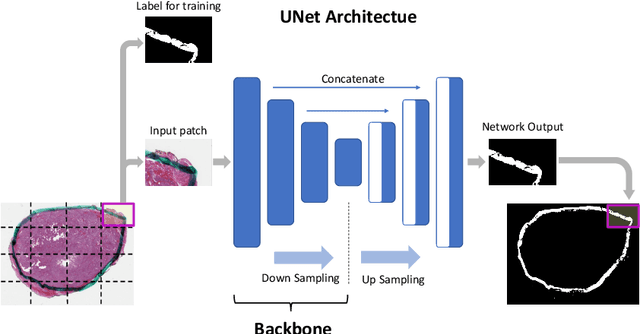
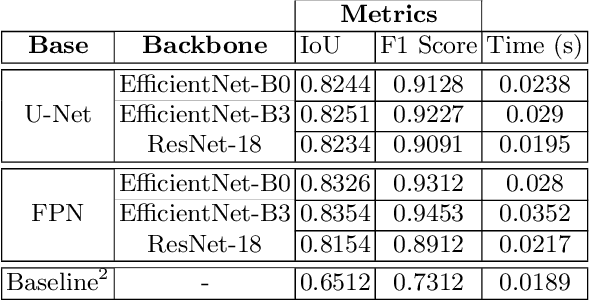
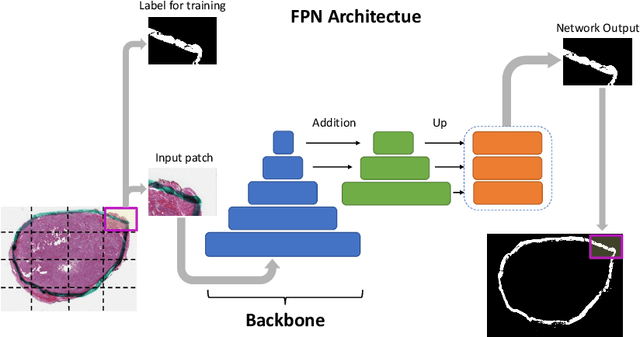
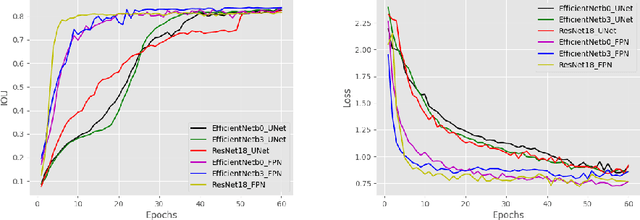
Abstract:Due to the recent advancements in machine vision, digital pathology has gained significant attention. Histopathology images are distinctly rich in visual information. The tissue glass slide images are utilized for disease diagnosis. Researchers study many methods to process histopathology images and facilitate fast and reliable diagnosis; therefore, the availability of high-quality slides becomes paramount. The quality of the images can be negatively affected when the glass slides are ink-marked by pathologists to delineate regions of interest. As an example, in one of the largest public histopathology datasets, The Cancer Genome Atlas (TCGA), approximately $12\%$ of the digitized slides are affected by manual delineations through ink markings. To process these open-access slide images and other repositories for the design and validation of new methods, an algorithm to detect the marked regions of the images is essential to avoid confusing tissue pixels with ink-colored pixels for computer methods. In this study, we propose to segment the ink-marked areas of pathology patches through a deep network. A dataset from $79$ whole slide images with $4,305$ patches was created and different networks were trained. Finally, the results showed an FPN model with the EffiecentNet-B3 as the backbone was found to be the superior configuration with an F1 score of $94.53\%$.
BlockCNN: A Deep Network for Artifact Removal and Image Compression
May 28, 2018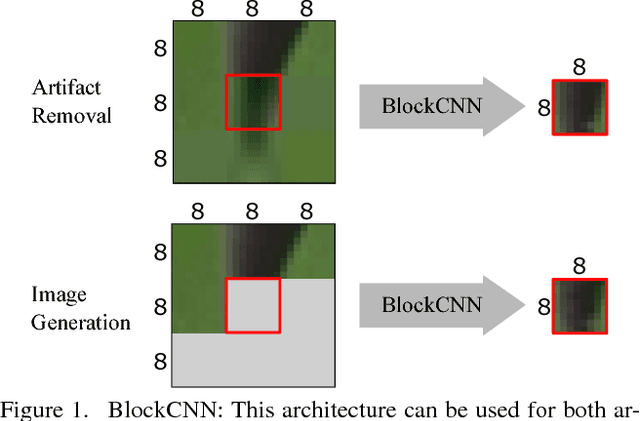
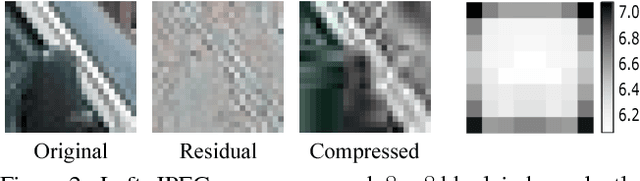
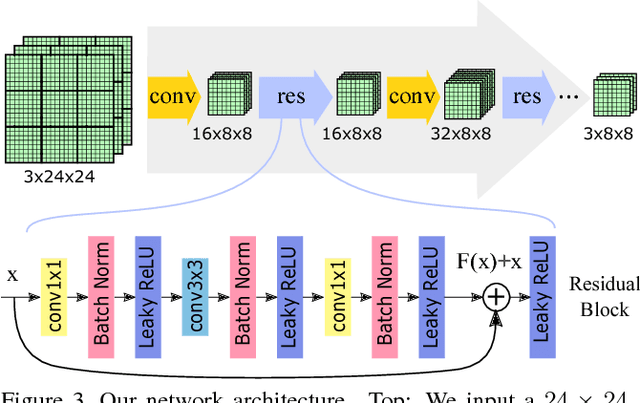
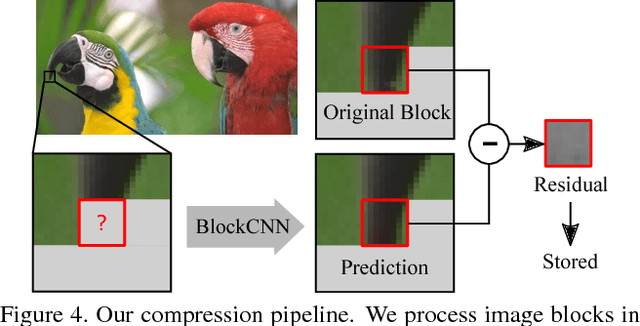
Abstract:We present a general technique that performs both artifact removal and image compression. For artifact removal, we input a JPEG image and try to remove its compression artifacts. For compression, we input an image and process its 8 by 8 blocks in a sequence. For each block, we first try to predict its intensities based on previous blocks; then, we store a residual with respect to the input image. Our technique reuses JPEG's legacy compression and decompression routines. Both our artifact removal and our image compression techniques use the same deep network, but with different training weights. Our technique is simple and fast and it significantly improves the performance of artifact removal and image compression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge