Manit Zaveri
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021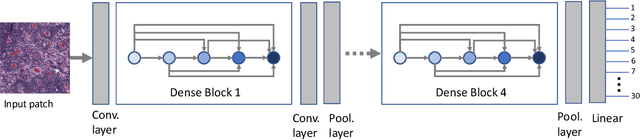
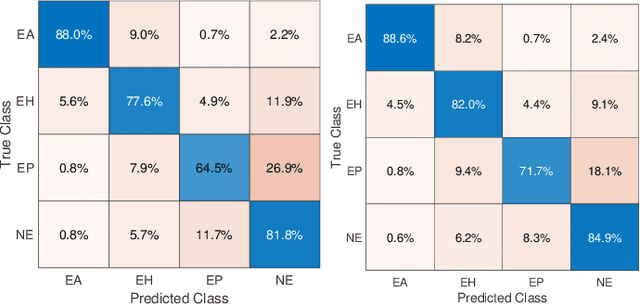
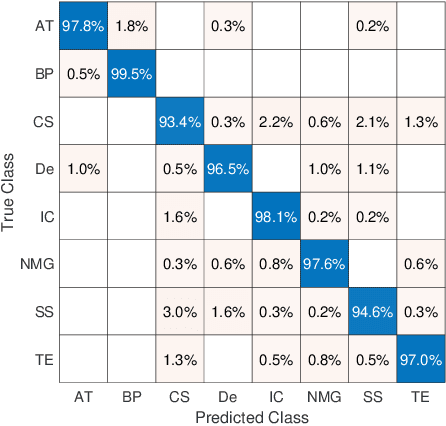
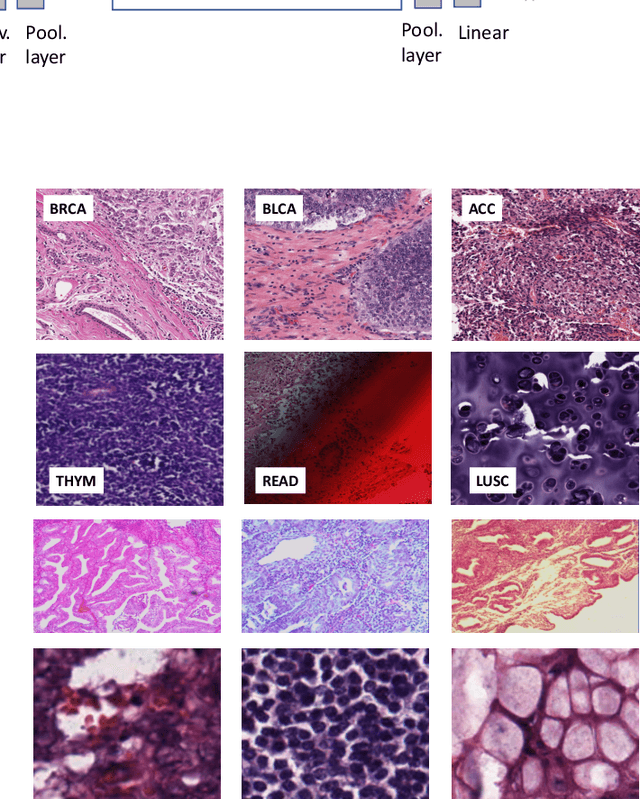
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Recognizing Magnification Levels in Microscopic Snapshots
May 07, 2020


Abstract:Recent advances in digital imaging has transformed computer vision and machine learning to new tools for analyzing pathology images. This trend could automate some of the tasks in the diagnostic pathology and elevate the pathologist workload. The final step of any cancer diagnosis procedure is performed by the expert pathologist. These experts use microscopes with high level of optical magnification to observe minute characteristics of the tissue acquired through biopsy and fixed on glass slides. Switching between different magnifications, and finding the magnification level at which they identify the presence or absence of malignant tissues is important. As the majority of pathologists still use light microscopy, compared to digital scanners, in many instance a mounted camera on the microscope is used to capture snapshots from significant field-of-views. Repositories of such snapshots usually do not contain the magnification information. In this paper, we extract deep features of the images available on TCGA dataset with known magnification to train a classifier for magnification recognition. We compared the results with LBP, a well-known handcrafted feature extraction method. The proposed approach achieved a mean accuracy of 96% when a multi-layer perceptron was trained as a classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge