Milad Sikaroudi
ALFA -- Leveraging All Levels of Feature Abstraction for Enhancing the Generalization of Histopathology Image Classification Across Unseen Hospitals
Aug 09, 2023Abstract:We propose an exhaustive methodology that leverages all levels of feature abstraction, targeting an enhancement in the generalizability of image classification to unobserved hospitals. Our approach incorporates augmentation-based self-supervision with common distribution shifts in histopathology scenarios serving as the pretext task. This enables us to derive invariant features from training images without relying on training labels, thereby covering different abstraction levels. Moving onto the subsequent abstraction level, we employ a domain alignment module to facilitate further extraction of invariant features across varying training hospitals. To represent the highly specific features of participating hospitals, an encoder is trained to classify hospital labels, independent of their diagnostic labels. The features from each of these encoders are subsequently disentangled to minimize redundancy and segregate the features. This representation, which spans a broad spectrum of semantic information, enables the development of a model demonstrating increased robustness to unseen images from disparate distributions. Experimental results from the PACS dataset (a domain generalization benchmark), a synthetic dataset created by applying histopathology-specific jitters to the MHIST dataset (defining different domains with varied distribution shifts), and a Renal Cell Carcinoma dataset derived from four image repositories from TCGA, collectively indicate that our proposed model is adept at managing varying levels of image granularity. Thus, it shows improved generalizability when faced with new, out-of-distribution hospital images.
Comments on 'Fast and scalable search of whole-slide images via self-supervised deep learning'
Apr 18, 2023Abstract:Chen et al. [Chen2022] recently published the article 'Fast and scalable search of whole-slide images via self-supervised deep learning' in Nature Biomedical Engineering. The authors call their method 'self-supervised image search for histology', short SISH. We express our concerns that SISH is an incremental modification of Yottixel, has used MinMax binarization but does not cite the original works, and is based on a misnomer 'self-supervised image search'. As well, we point to several other concerns regarding experiments and comparisons performed by Chen et al.
Cluster Based Secure Multi-Party Computation in Federated Learning for Histopathology Images
Aug 21, 2022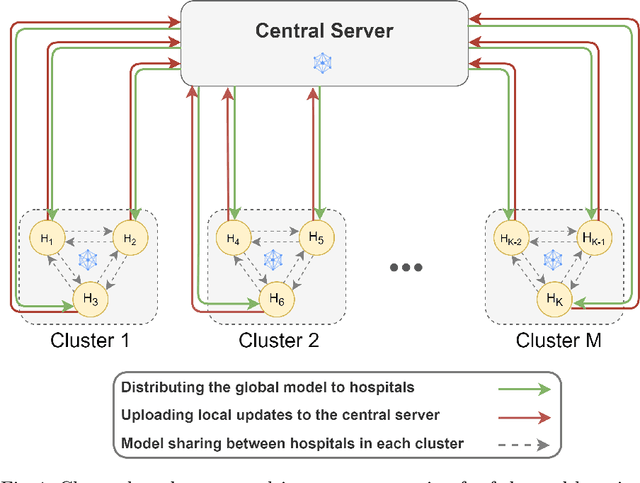
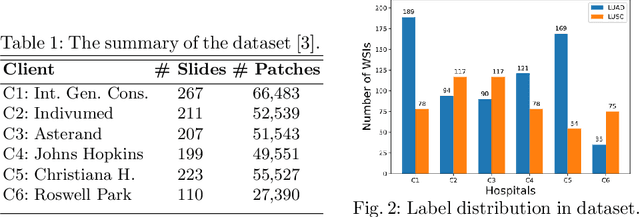
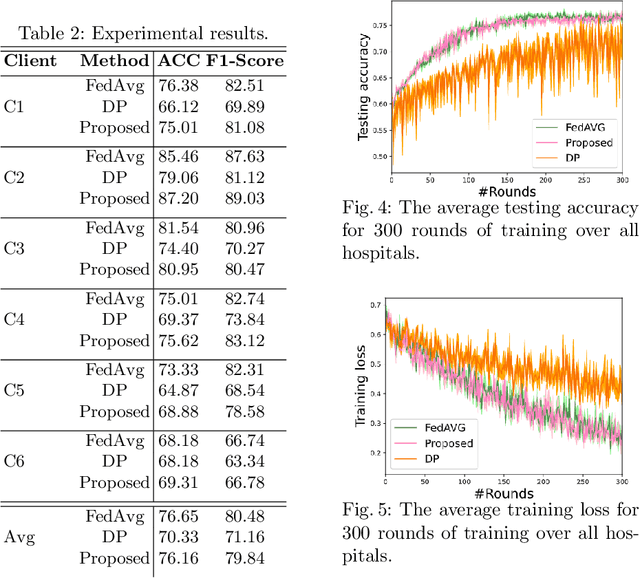
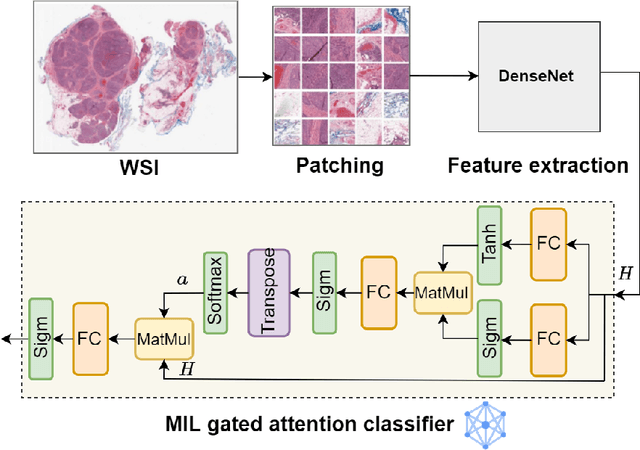
Abstract:Federated learning (FL) is a decentralized method enabling hospitals to collaboratively learn a model without sharing private patient data for training. In FL, participant hospitals periodically exchange training results rather than training samples with a central server. However, having access to model parameters or gradients can expose private training data samples. To address this challenge, we adopt secure multiparty computation (SMC) to establish a privacy-preserving federated learning framework. In our proposed method, the hospitals are divided into clusters. After local training, each hospital splits its model weights among other hospitals in the same cluster such that no single hospital can retrieve other hospitals' weights on its own. Then, all hospitals sum up the received weights, sending the results to the central server. Finally, the central server aggregates the results, retrieving the average of models' weights and updating the model without having access to individual hospitals' weights. We conduct experiments on a publicly available repository, The Cancer Genome Atlas (TCGA). We compare the performance of the proposed framework with differential privacy and federated averaging as the baseline. The results reveal that compared to differential privacy, our framework can achieve higher accuracy with no privacy leakage risk at a cost of higher communication overhead.
Hospital-Agnostic Image Representation Learning in Digital Pathology
Apr 05, 2022

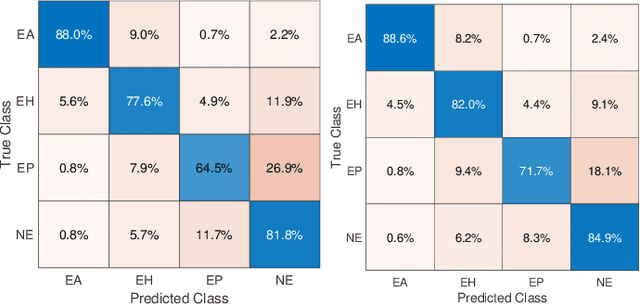
Abstract:Whole Slide Images (WSIs) in digital pathology are used to diagnose cancer subtypes. The difference in procedures to acquire WSIs at various trial sites gives rise to variability in the histopathology images, thus making consistent diagnosis challenging. These differences may stem from variability in image acquisition through multi-vendor scanners, variable acquisition parameters, and differences in staining procedure; as well, patient demographics may bias the glass slide batches before image acquisition. These variabilities are assumed to cause a domain shift in the images of different hospitals. It is crucial to overcome this domain shift because an ideal machine-learning model must be able to work on the diverse sources of images, independent of the acquisition center. A domain generalization technique is leveraged in this study to improve the generalization capability of a Deep Neural Network (DNN), to an unseen histopathology image set (i.e., from an unseen hospital/trial site) in the presence of domain shift. According to experimental results, the conventional supervised-learning regime generalizes poorly to data collected from different hospitals. However, the proposed hospital-agnostic learning can improve the generalization considering the low-dimensional latent space representation visualization, and classification accuracy results.
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021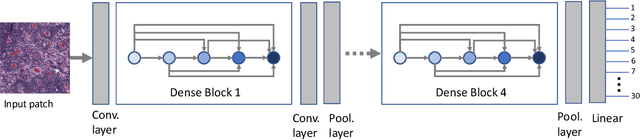
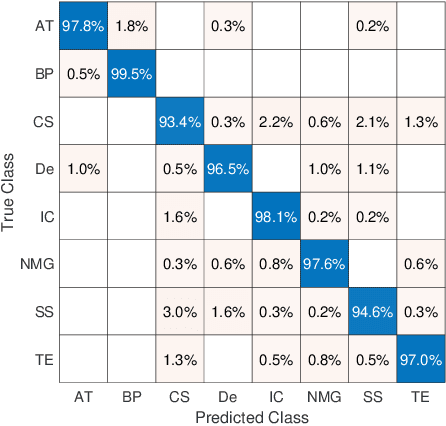
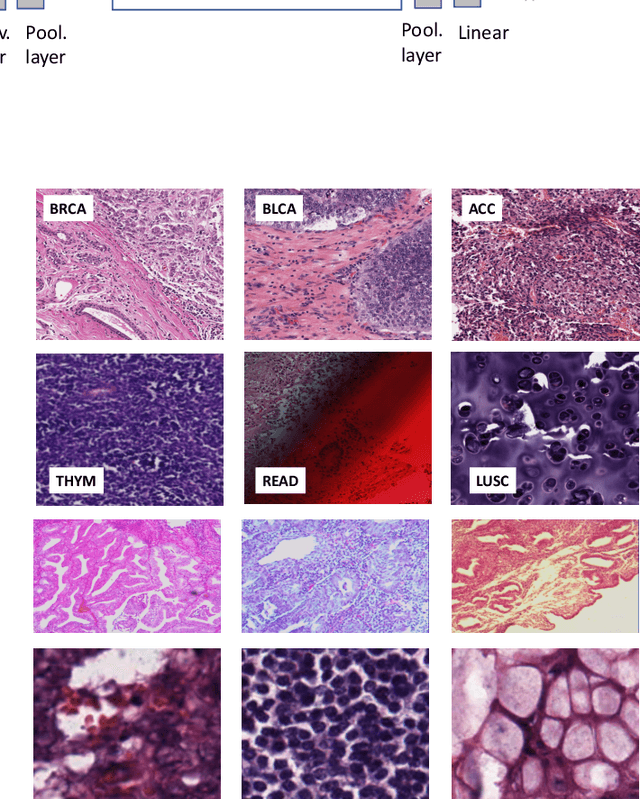
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
Magnification Generalization for Histopathology Image Embedding
Jan 18, 2021

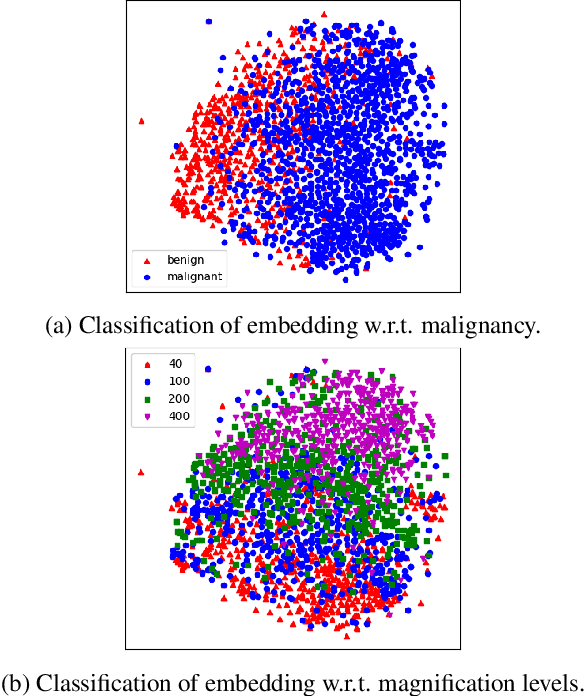

Abstract:Histopathology image embedding is an active research area in computer vision. Most of the embedding models exclusively concentrate on a specific magnification level. However, a useful task in histopathology embedding is to train an embedding space regardless of the magnification level. Two main approaches for tackling this goal are domain adaptation and domain generalization, where the target magnification levels may or may not be introduced to the model in training, respectively. Although magnification adaptation is a well-studied topic in the literature, this paper, to the best of our knowledge, is the first work on magnification generalization for histopathology image embedding. We use an episodic trainable domain generalization technique for magnification generalization, namely Model Agnostic Learning of Semantic Features (MASF), which works based on the Model Agnostic Meta-Learning (MAML) concept. Our experimental results on a breast cancer histopathology dataset with four different magnification levels show the proposed method's effectiveness for magnification generalization.
Batch-Incremental Triplet Sampling for Training Triplet Networks Using Bayesian Updating Theorem
Jul 10, 2020



Abstract:Variants of Triplet networks are robust entities for learning a discriminative embedding subspace. There exist different triplet mining approaches for selecting the most suitable training triplets. Some of these mining methods rely on the extreme distances between instances, and some others make use of sampling. However, sampling from stochastic distributions of data rather than sampling merely from the existing embedding instances can provide more discriminative information. In this work, we sample triplets from distributions of data rather than from existing instances. We consider a multivariate normal distribution for the embedding of each class. Using Bayesian updating and conjugate priors, we update the distributions of classes dynamically by receiving the new mini-batches of training data. The proposed triplet mining with Bayesian updating can be used with any triplet-based loss function, e.g., triplet-loss or Neighborhood Component Analysis (NCA) loss. Accordingly, Our triplet mining approaches are called Bayesian Updating Triplet (BUT) and Bayesian Updating NCA (BUNCA), depending on which loss function is being used. Experimental results on two public datasets, namely MNIST and histopathology colorectal cancer (CRC), substantiate the effectiveness of the proposed triplet mining method.
Offline versus Online Triplet Mining based on Extreme Distances of Histopathology Patches
Jul 04, 2020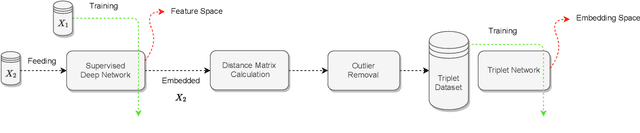
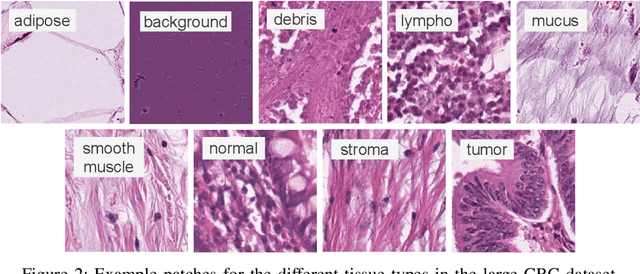
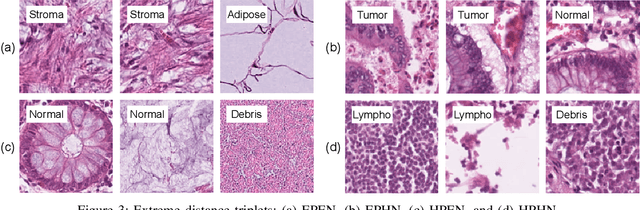
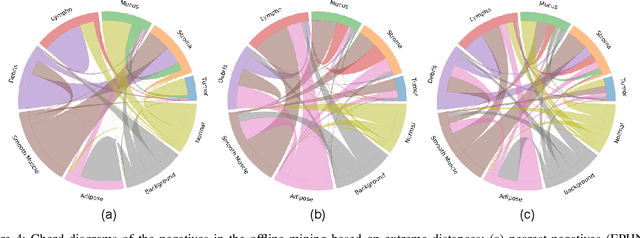
Abstract:We analyze the effect of offline and online triplet mining for colorectal cancer (CRC) histopathology dataset containing 100,000 patches. We consider the extreme, i.e., farthest and nearest patches with respect to a given anchor, both in online and offline mining. While many works focus solely on how to select the triplets online (batch-wise), we also study the effect of extreme distances and neighbor patches before training in an offline fashion. We analyze the impacts of extreme cases for offline versus online mining, including easy positive, batch semi-hard, and batch hard triplet mining as well as the neighborhood component analysis loss, its proxy version, and distance weighted sampling. We also investigate online approaches based on extreme distance and comprehensively compare the performance of offline and online mining based on the data patterns and explain offline mining as a tractable generalization of the online mining with large mini-batch size. As well, we discuss the relations of different colorectal tissue types in terms of extreme distances. We found that offline mining can generate a better statistical representation of the population by working on the whole dataset.
Supervision and Source Domain Impact on Representation Learning: A Histopathology Case Study
May 10, 2020



Abstract:As many algorithms depend on a suitable representation of data, learning unique features is considered a crucial task. Although supervised techniques using deep neural networks have boosted the performance of representation learning, the need for a large set of labeled data limits the application of such methods. As an example, high-quality delineations of regions of interest in the field of pathology is a tedious and time-consuming task due to the large image dimensions. In this work, we explored the performance of a deep neural network and triplet loss in the area of representation learning. We investigated the notion of similarity and dissimilarity in pathology whole-slide images and compared different setups from unsupervised and semi-supervised to supervised learning in our experiments. Additionally, different approaches were tested, applying few-shot learning on two publicly available pathology image datasets. We achieved high accuracy and generalization when the learned representations were applied to two different pathology datasets.
Fisher Discriminant Triplet and Contrastive Losses for Training Siamese Networks
Apr 05, 2020



Abstract:Siamese neural network is a very powerful architecture for both feature extraction and metric learning. It usually consists of several networks that share weights. The Siamese concept is topology-agnostic and can use any neural network as its backbone. The two most popular loss functions for training these networks are the triplet and contrastive loss functions. In this paper, we propose two novel loss functions, named Fisher Discriminant Triplet (FDT) and Fisher Discriminant Contrastive (FDC). The former uses anchor-neighbor-distant triplets while the latter utilizes pairs of anchor-neighbor and anchor-distant samples. The FDT and FDC loss functions are designed based on the statistical formulation of the Fisher Discriminant Analysis (FDA), which is a linear subspace learning method. Our experiments on the MNIST and two challenging and publicly available histopathology datasets show the effectiveness of the proposed loss functions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge