Morteza Babaei
Cluster Based Secure Multi-Party Computation in Federated Learning for Histopathology Images
Aug 21, 2022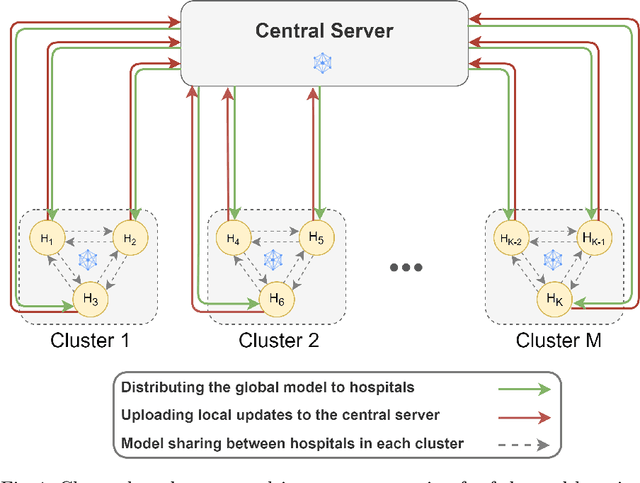
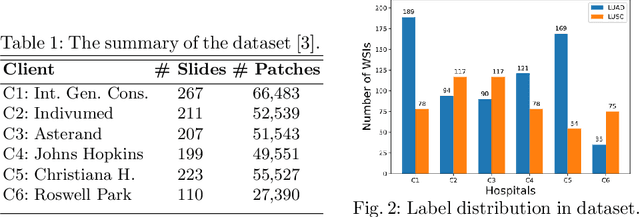
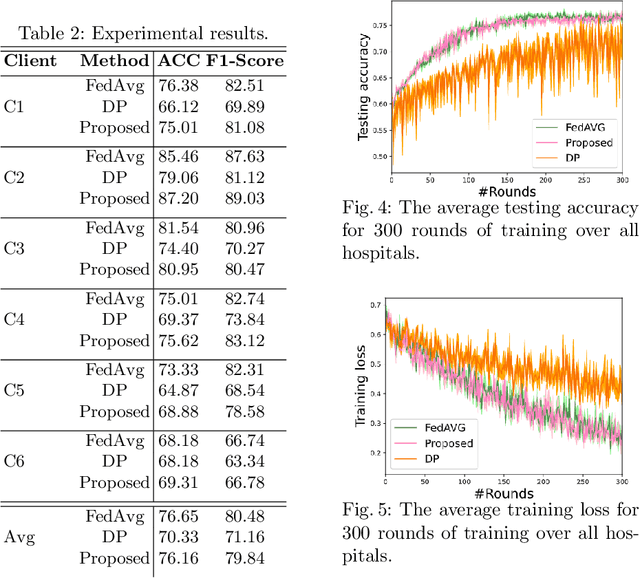
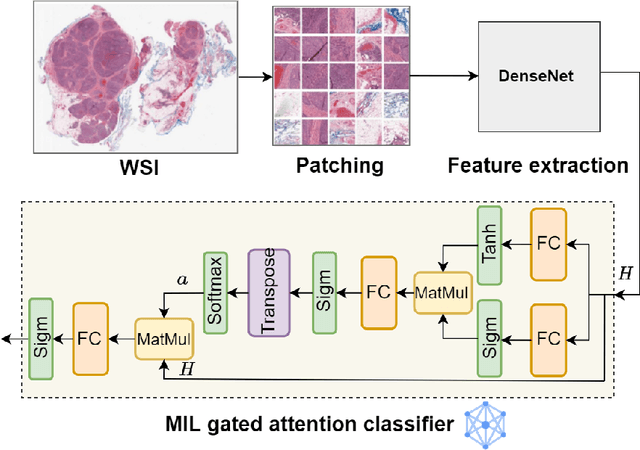
Abstract:Federated learning (FL) is a decentralized method enabling hospitals to collaboratively learn a model without sharing private patient data for training. In FL, participant hospitals periodically exchange training results rather than training samples with a central server. However, having access to model parameters or gradients can expose private training data samples. To address this challenge, we adopt secure multiparty computation (SMC) to establish a privacy-preserving federated learning framework. In our proposed method, the hospitals are divided into clusters. After local training, each hospital splits its model weights among other hospitals in the same cluster such that no single hospital can retrieve other hospitals' weights on its own. Then, all hospitals sum up the received weights, sending the results to the central server. Finally, the central server aggregates the results, retrieving the average of models' weights and updating the model without having access to individual hospitals' weights. We conduct experiments on a publicly available repository, The Cancer Genome Atlas (TCGA). We compare the performance of the proposed framework with differential privacy and federated averaging as the baseline. The results reveal that compared to differential privacy, our framework can achieve higher accuracy with no privacy leakage risk at a cost of higher communication overhead.
A Comparative Study of U-Net Topologies for Background Removal in Histopathology Images
Jun 08, 2020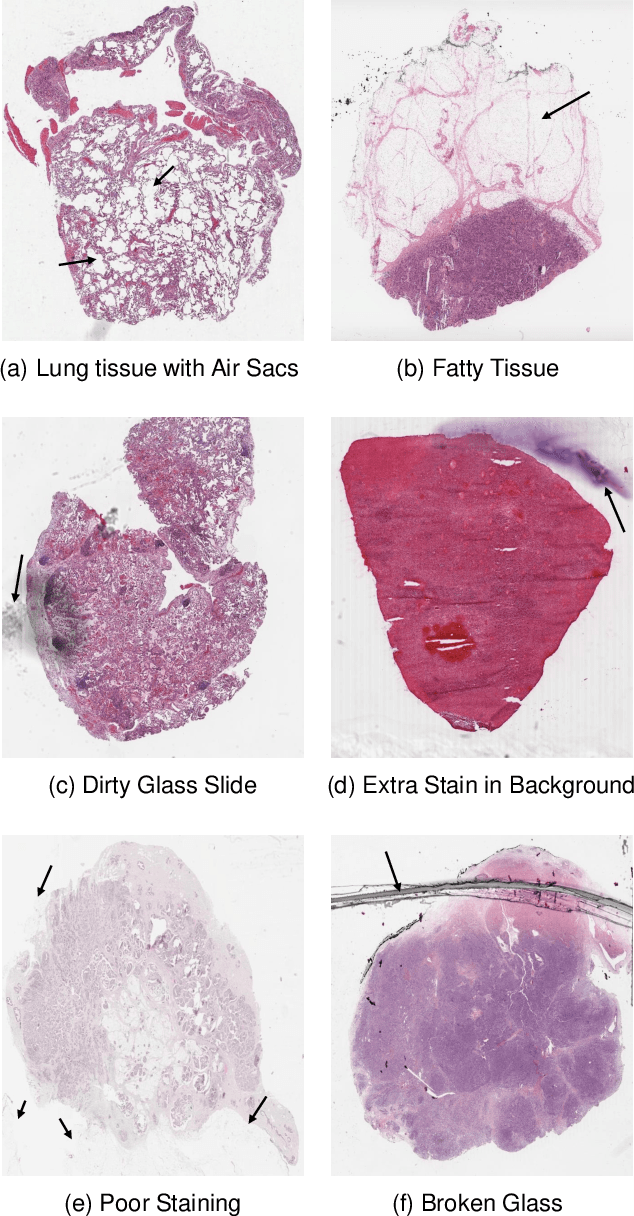
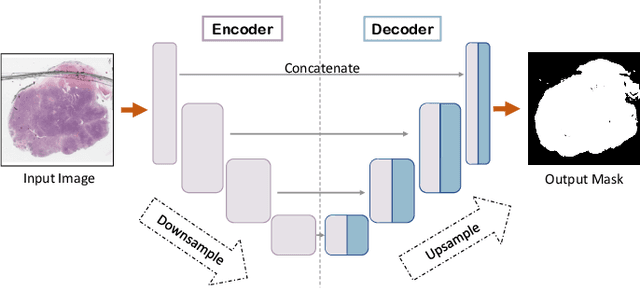
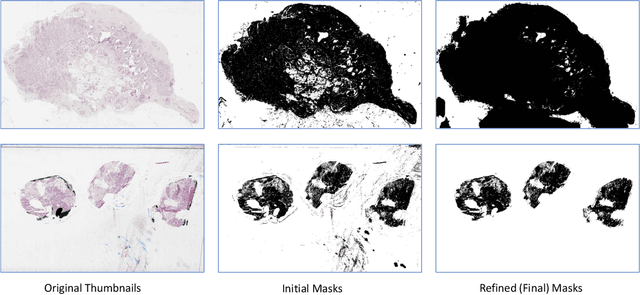
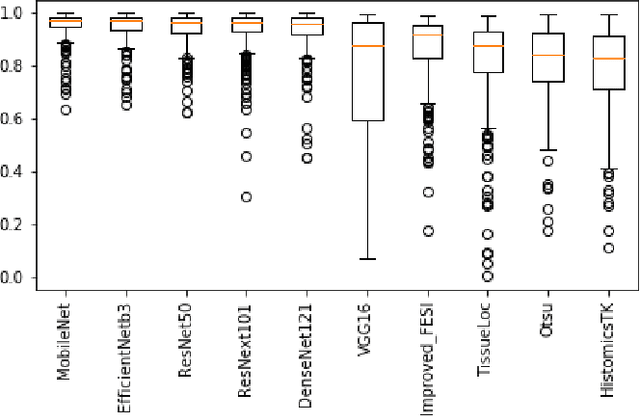
Abstract:During the last decade, the digitization of pathology has gained considerable momentum. Digital pathology offers many advantages including more efficient workflows, easier collaboration as well as a powerful venue for telepathology. At the same time, applying Computer-Aided Diagnosis (CAD) on Whole Slide Images (WSIs) has received substantial attention as a direct result of the digitization. The first step in any image analysis is to extract the tissue. Hence, background removal is an essential prerequisite for efficient and accurate results for many algorithms. In spite of the obvious discrimination for human operators, the identification of tissue regions in WSIs could be challenging for computers, mainly due to the existence of color variations and artifacts. Moreover, some cases such as alveolar tissue types, fatty tissues, and tissues with poor staining are difficult to detect. In this paper, we perform experiments on U-Net architecture with different network backbones (different topologies) to remove the background as well as artifacts from WSIs in order to extract the tissue regions. We compare a wide range of backbone networks including MobileNet, VGG16, EfficientNet-B3, ResNet50, ResNext101 and DenseNet121. We trained and evaluated the network on a manually labeled subset of The Cancer Genome Atlas (TCGA) Dataset. EfficientNet-B3 and MobileNet by almost 99% sensitivity and specificity reached the best results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge