Maral Rasoolijaberi
Fine-Tuning and Training of DenseNet for Histopathology Image Representation Using TCGA Diagnostic Slides
Jan 20, 2021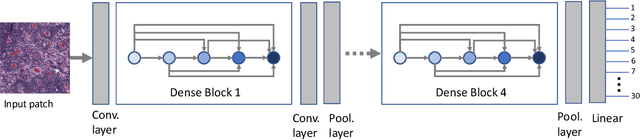
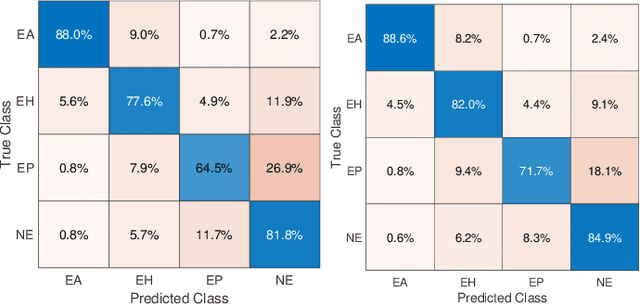
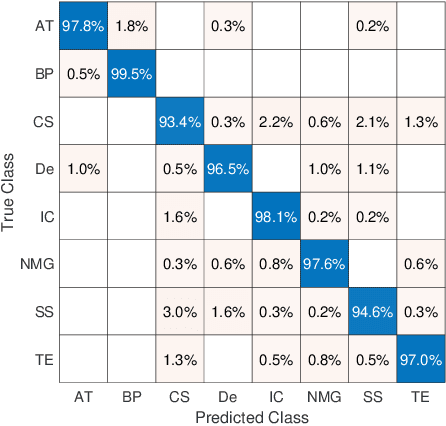
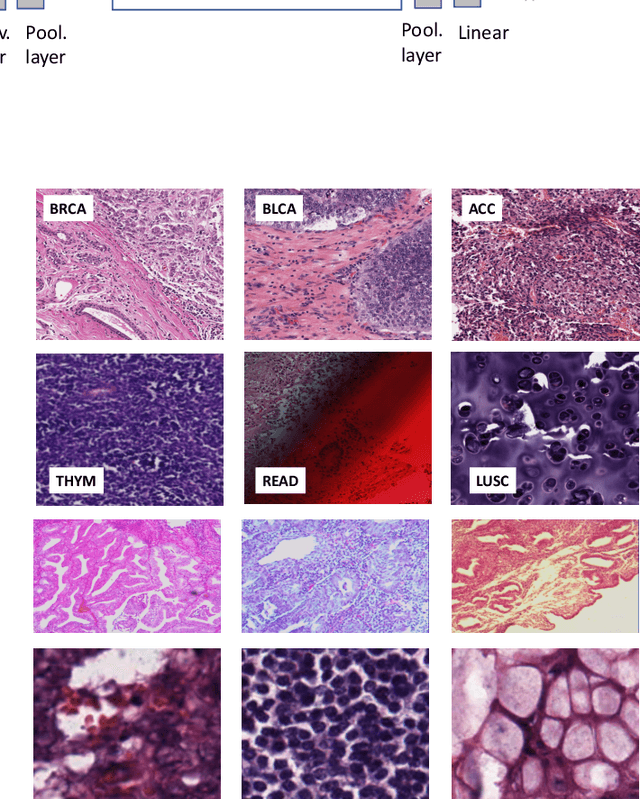
Abstract:Feature vectors provided by pre-trained deep artificial neural networks have become a dominant source for image representation in recent literature. Their contribution to the performance of image analysis can be improved through finetuning. As an ultimate solution, one might even train a deep network from scratch with the domain-relevant images, a highly desirable option which is generally impeded in pathology by lack of labeled images and the computational expense. In this study, we propose a new network, namely KimiaNet, that employs the topology of the DenseNet with four dense blocks, fine-tuned and trained with histopathology images in different configurations. We used more than 240,000 image patches with 1000x1000 pixels acquired at 20x magnification through our proposed "highcellularity mosaic" approach to enable the usage of weak labels of 7,126 whole slide images of formalin-fixed paraffin-embedded human pathology samples publicly available through the The Cancer Genome Atlas (TCGA) repository. We tested KimiaNet using three public datasets, namely TCGA, endometrial cancer images, and colorectal cancer images by evaluating the performance of search and classification when corresponding features of different networks are used for image representation. As well, we designed and trained multiple convolutional batch-normalized ReLU (CBR) networks. The results show that KimiaNet provides superior results compared to the original DenseNet and smaller CBR networks when used as feature extractor to represent histopathology images.
A Comparative Study of U-Net Topologies for Background Removal in Histopathology Images
Jun 08, 2020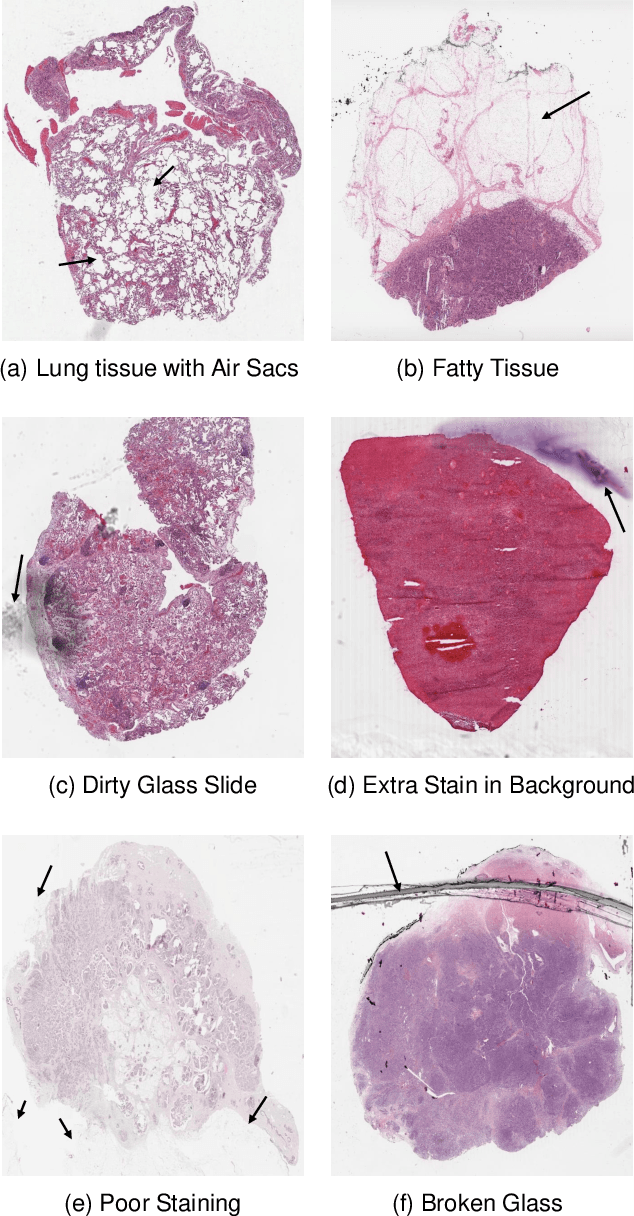
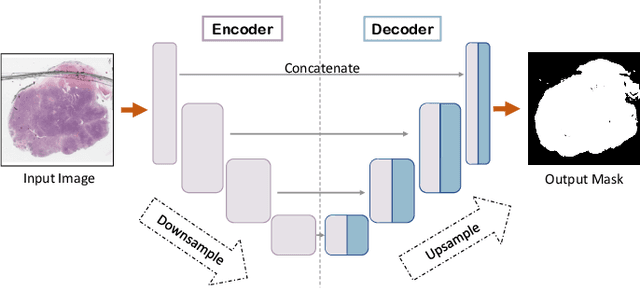
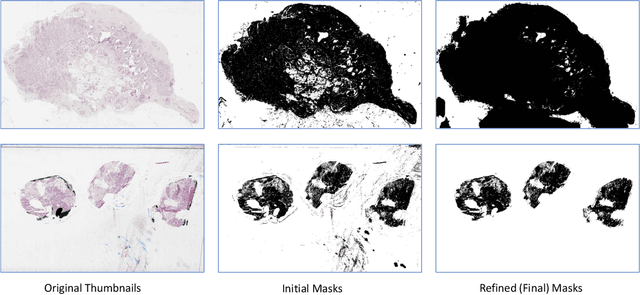
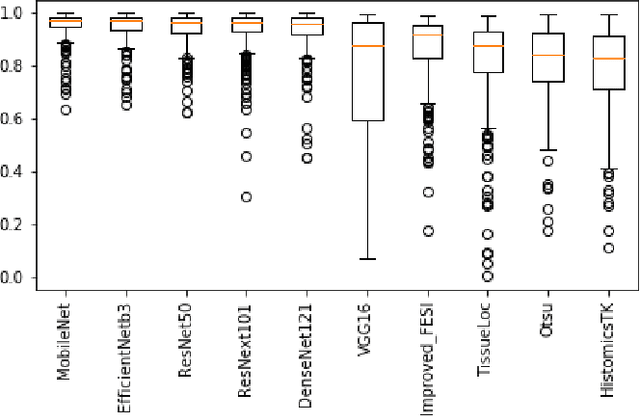
Abstract:During the last decade, the digitization of pathology has gained considerable momentum. Digital pathology offers many advantages including more efficient workflows, easier collaboration as well as a powerful venue for telepathology. At the same time, applying Computer-Aided Diagnosis (CAD) on Whole Slide Images (WSIs) has received substantial attention as a direct result of the digitization. The first step in any image analysis is to extract the tissue. Hence, background removal is an essential prerequisite for efficient and accurate results for many algorithms. In spite of the obvious discrimination for human operators, the identification of tissue regions in WSIs could be challenging for computers, mainly due to the existence of color variations and artifacts. Moreover, some cases such as alveolar tissue types, fatty tissues, and tissues with poor staining are difficult to detect. In this paper, we perform experiments on U-Net architecture with different network backbones (different topologies) to remove the background as well as artifacts from WSIs in order to extract the tissue regions. We compare a wide range of backbone networks including MobileNet, VGG16, EfficientNet-B3, ResNet50, ResNext101 and DenseNet121. We trained and evaluated the network on a manually labeled subset of The Cancer Genome Atlas (TCGA) Dataset. EfficientNet-B3 and MobileNet by almost 99% sensitivity and specificity reached the best results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge