Adrian Batten
Department of Pathology, Grand River Hospital, Kitchener, ON, Canada., and
Composite Biomarker Image for Advanced Visualization in Histopathology
Apr 24, 2023



Abstract:Immunohistochemistry (IHC) biomarkers are essential tools for reliable cancer diagnosis and subtyping. It requires cross-staining comparison among Whole Slide Images (WSIs) of IHCs and hematoxylin and eosin (H&E) slides. Currently, pathologists examine the visually co-localized areas across IHC and H&E glass slides for a final diagnosis, which is a tedious and challenging task. Moreover, visually inspecting different IHC slides back and forth to analyze local co-expressions is inherently subjective and prone to error, even when carried out by experienced pathologists. Relying on digital pathology, we propose Composite Biomarker Image (CBI) in this work. CBI is a single image that can be composed using different filtered IHC biomarker images for better visualization. We present a CBI image produced in two steps by the proposed solution for better visualization and hence more efficient clinical workflow. In the first step, IHC biomarker images are aligned with the H&E images using one coordinate system and orientation. In the second step, the positive or negative IHC regions from each biomarker image (based on the pathologists recommendation) are filtered and combined into one image using a fuzzy inference system. For evaluation, the resulting CBI images, from the proposed system, were evaluated qualitatively by the expert pathologists. The CBI concept helps the pathologists to identify the suspected target tissues more easily, which could be further assessed by examining the actual WSIs at the same suspected regions.
Automatic Multi-Stain Registration of Whole Slide Images in Histopathology
Jul 29, 2021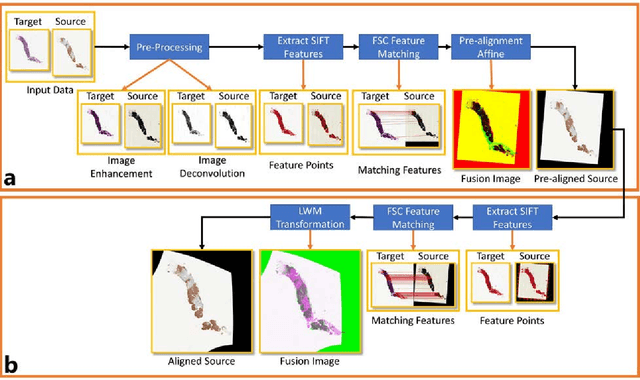
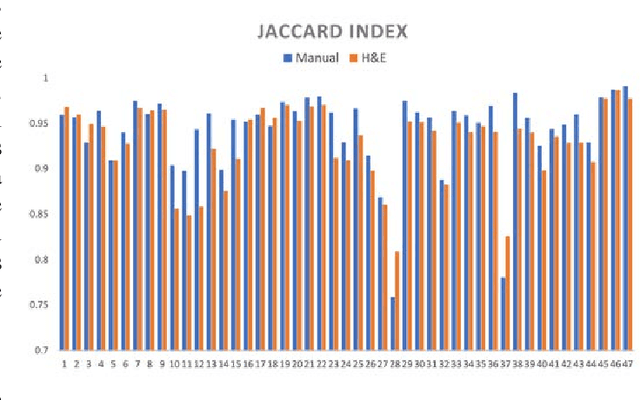
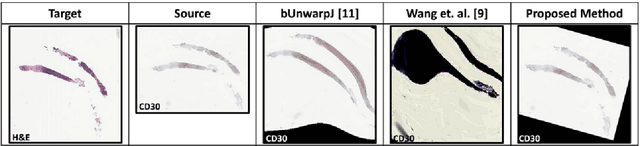
Abstract:Joint analysis of multiple biomarker images and tissue morphology is important for disease diagnosis, treatment planning and drug development. It requires cross-staining comparison among Whole Slide Images (WSIs) of immuno-histochemical and hematoxylin and eosin (H&E) microscopic slides. However, automatic, and fast cross-staining alignment of enormous gigapixel WSIs at single-cell precision is challenging. In addition to morphological deformations introduced during slide preparation, there are large variations in cell appearance and tissue morphology across different staining. In this paper, we propose a two-step automatic feature-based cross-staining WSI alignment to assist localization of even tiny metastatic foci in the assessment of lymph node. Image pairs were aligned allowing for translation, rotation, and scaling. The registration was performed automatically by first detecting landmarks in both images, using the scale-invariant image transform (SIFT), followed by the fast sample consensus (FSC) protocol for finding point correspondences and finally aligned the images. The Registration results were evaluated using both visual and quantitative criteria using the Jaccard index. The average Jaccard similarity index of the results produced by the proposed system is 0.942 when compared with the manual registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge