Shuxin Yang
PolSAM: Polarimetric Scattering Mechanism Informed Segment Anything Model
Dec 17, 2024Abstract:PolSAR data presents unique challenges due to its rich and complex characteristics. Existing data representations, such as complex-valued data, polarimetric features, and amplitude images, are widely used. However, these formats often face issues related to usability, interpretability, and data integrity. Most feature extraction networks for PolSAR are small, limiting their ability to capture features effectively. To address these issues, We propose the Polarimetric Scattering Mechanism-Informed SAM (PolSAM), an enhanced Segment Anything Model (SAM) that integrates domain-specific scattering characteristics and a novel prompt generation strategy. PolSAM introduces Microwave Vision Data (MVD), a lightweight and interpretable data representation derived from polarimetric decomposition and semantic correlations. We propose two key components: the Feature-Level Fusion Prompt (FFP), which fuses visual tokens from pseudo-colored SAR images and MVD to address modality incompatibility in the frozen SAM encoder, and the Semantic-Level Fusion Prompt (SFP), which refines sparse and dense segmentation prompts using semantic information. Experimental results on the PhySAR-Seg datasets demonstrate that PolSAM significantly outperforms existing SAM-based and multimodal fusion models, improving segmentation accuracy, reducing data storage, and accelerating inference time. The source code and datasets will be made publicly available at \url{https://github.com/XAI4SAR/PolSAM}.
Physics-Guided Detector for SAR Airplanes
Nov 19, 2024Abstract:The disperse structure distributions (discreteness) and variant scattering characteristics (variability) of SAR airplane targets lead to special challenges of object detection and recognition. The current deep learning-based detectors encounter challenges in distinguishing fine-grained SAR airplanes against complex backgrounds. To address it, we propose a novel physics-guided detector (PGD) learning paradigm for SAR airplanes that comprehensively investigate their discreteness and variability to improve the detection performance. It is a general learning paradigm that can be extended to different existing deep learning-based detectors with "backbone-neck-head" architectures. The main contributions of PGD include the physics-guided self-supervised learning, feature enhancement, and instance perception, denoted as PGSSL, PGFE, and PGIP, respectively. PGSSL aims to construct a self-supervised learning task based on a wide range of SAR airplane targets that encodes the prior knowledge of various discrete structure distributions into the embedded space. Then, PGFE enhances the multi-scale feature representation of a detector, guided by the physics-aware information learned from PGSSL. PGIP is constructed at the detection head to learn the refined and dominant scattering point of each SAR airplane instance, thus alleviating the interference from the complex background. We propose two implementations, denoted as PGD and PGD-Lite, and apply them to various existing detectors with different backbones and detection heads. The experiments demonstrate the flexibility and effectiveness of the proposed PGD, which can improve existing detectors on SAR airplane detection with fine-grained classification task (an improvement of 3.1\% mAP most), and achieve the state-of-the-art performance (90.7\% mAP) on SAR-AIRcraft-1.0 dataset. The project is open-source at \url{https://github.com/XAI4SAR/PGD}.
OCC-MLLM-Alpha:Empowering Multi-modal Large Language Model for the Understanding of Occluded Objects with Self-Supervised Test-Time Learning
Oct 02, 2024



Abstract:There is a gap in the understanding of occluded objects in existing large-scale visual language multi-modal models. Current state-of-the-art multi-modal models fail to provide satisfactory results in describing occluded objects through universal visual encoders and supervised learning strategies. Therefore, we introduce a multi-modal large language framework and corresponding self-supervised learning strategy with support of 3D generation. We start our experiments comparing with the state-of-the-art models in the evaluation of a large-scale dataset SOMVideo [18]. The initial results demonstrate the improvement of 16.92% in comparison with the state-of-the-art VLM models.
Universal Segmentation of 33 Anatomies
Mar 04, 2022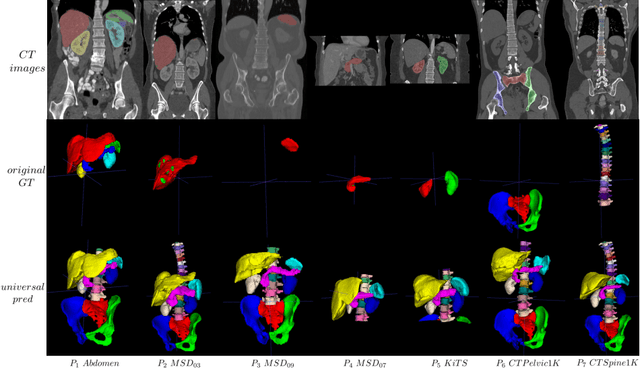
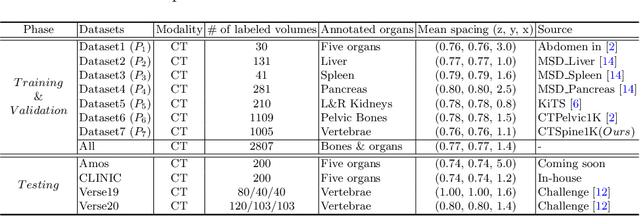
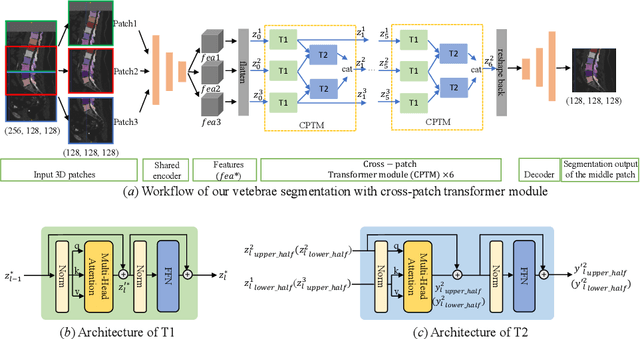
Abstract:In the paper, we present an approach for learning a single model that universally segments 33 anatomical structures, including vertebrae, pelvic bones, and abdominal organs. Our model building has to address the following challenges. Firstly, while it is ideal to learn such a model from a large-scale, fully-annotated dataset, it is practically hard to curate such a dataset. Thus, we resort to learn from a union of multiple datasets, with each dataset containing the images that are partially labeled. Secondly, along the line of partial labelling, we contribute an open-source, large-scale vertebra segmentation dataset for the benefit of spine analysis community, CTSpine1K, boasting over 1,000 3D volumes and over 11K annotated vertebrae. Thirdly, in a 3D medical image segmentation task, due to the limitation of GPU memory, we always train a model using cropped patches as inputs instead a whole 3D volume, which limits the amount of contextual information to be learned. To this, we propose a cross-patch transformer module to fuse more information in adjacent patches, which enlarges the aggregated receptive field for improved segmentation performance. This is especially important for segmenting, say, the elongated spine. Based on 7 partially labeled datasets that collectively contain about 2,800 3D volumes, we successfully learn such a universal model. Finally, we evaluate the universal model on multiple open-source datasets, proving that our model has a good generalization performance and can potentially serve as a solid foundation for downstream tasks.
Radiology Report Generation with a Learned Knowledge Base and Multi-modal Alignment
Dec 30, 2021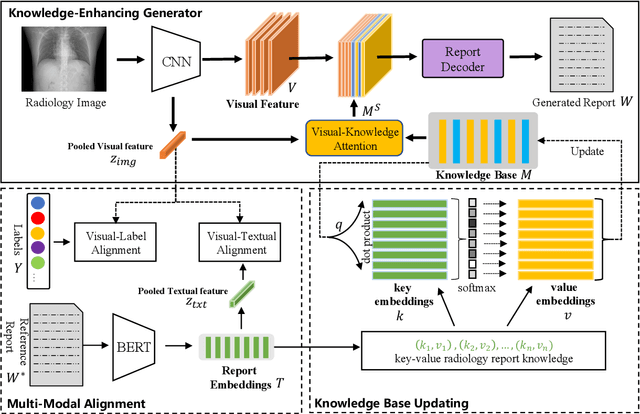
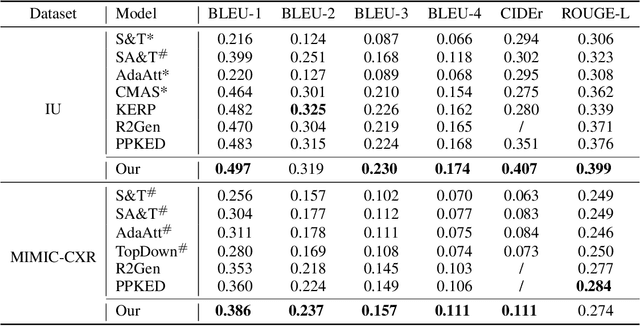
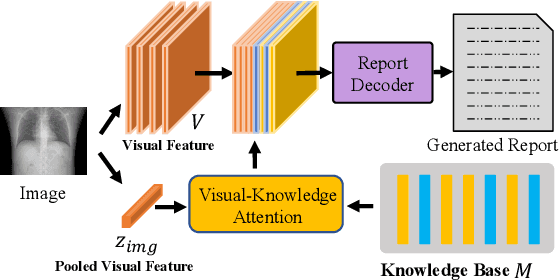
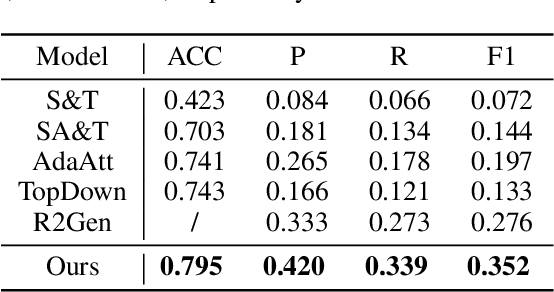
Abstract:In clinics, a radiology report is crucial for guiding a patient's treatment. Unfortunately, report writing imposes a heavy burden on radiologists. To effectively reduce such a burden, we hereby present an automatic, multi-modal approach for report generation from chest x-ray. Our approach, motivated by the observation that the descriptions in radiology reports are highly correlated with the x-ray images, features two distinct modules: (i) Learned knowledge base. To absorb the knowledge embedded in the above-mentioned correlation, we automatically build a knowledge base based on textual embedding. (ii) Multi-modal alignment. To promote the semantic alignment among reports, disease labels and images, we explicitly utilize textual embedding to guide the learning of the visual feature space. We evaluate the performance of the proposed model using metrics from both natural language generation and clinic efficacy on the public IU and MIMIC-CXR datasets. Our ablation study shows that each module contributes to improving the quality of generated reports. Furthermore, with the aid of both modules, our approach clearly outperforms state-of-the-art methods.
Knowledge Matters: Radiology Report Generation with General and Specific Knowledge
Dec 30, 2021



Abstract:Automatic radiology report generation is critical in clinics which can relieve experienced radiologists from the heavy workload and remind inexperienced radiologists of misdiagnosis or missed diagnose. Existing approaches mainly formulate radiology report generation as an image captioning task and adopt the encoder-decoder framework. However, in the medical domain, such pure data-driven approaches suffer from the following problems: 1) visual and textual bias problem; 2) lack of expert knowledge. In this paper, we propose a knowledge-enhanced radiology report generation approach introduces two types of medical knowledge: 1) General knowledge, which is input independent and provides the broad knowledge for report generation; 2) Specific knowledge, which is input dependent and provides the fine-grained knowledge for report generation. To fully utilize both the general and specific knowledge, we also propose a knowledge-enhanced multi-head attention mechanism. By merging the visual features of the radiology image with general knowledge and specific knowledge, the proposed model can improve the quality of generated reports. Experimental results on two publicly available datasets IU-Xray and MIMIC-CXR show that the proposed knowledge enhanced approach outperforms state-of-the-art image captioning based methods. Ablation studies also demonstrate that both general and specific knowledge can help to improve the performance of radiology report generation.
CTSpine1K: A Large-Scale Dataset for Spinal Vertebrae Segmentation in Computed Tomography
Jun 10, 2021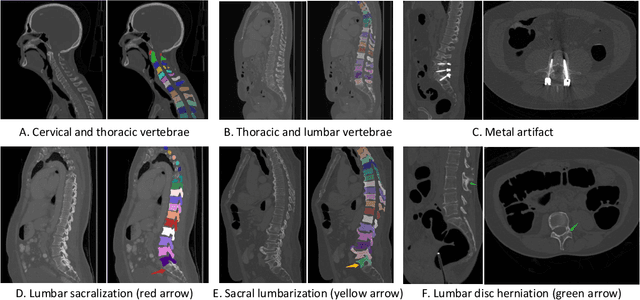
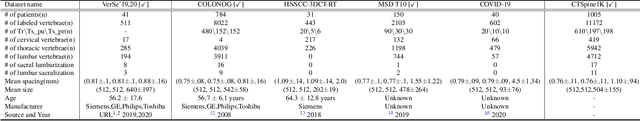
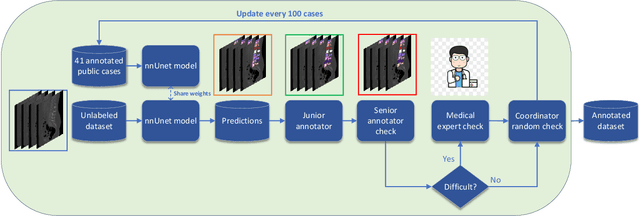
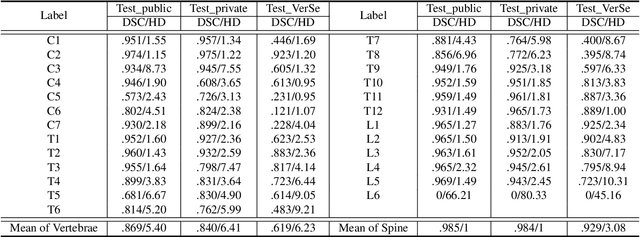
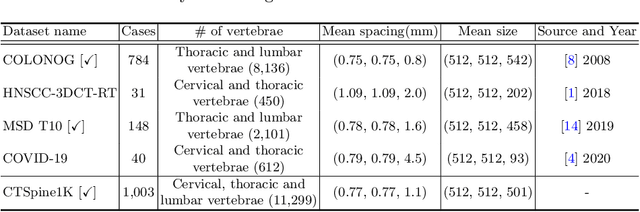
Abstract:Spine-related diseases have high morbidity and cause a huge burden of social cost. Spine imaging is an essential tool for noninvasively visualizing and assessing spinal pathology. Segmenting vertebrae in computed tomography (CT) images is the basis of quantitative medical image analysis for clinical diagnosis and surgery planning of spine diseases. Current publicly available annotated datasets on spinal vertebrae are small in size. Due to the lack of a large-scale annotated spine image dataset, the mainstream deep learning-based segmentation methods, which are data-driven, are heavily restricted. In this paper, we introduce a large-scale spine CT dataset, called CTSpine1K, curated from multiple sources for vertebra segmentation, which contains 1,005 CT volumes with over 11,100 labeled vertebrae belonging to different spinal conditions. Based on this dataset, we conduct several spinal vertebrae segmentation experiments to set the first benchmark. We believe that this large-scale dataset will facilitate further research in many spine-related image analysis tasks, including but not limited to vertebrae segmentation, labeling, 3D spine reconstruction from biplanar radiographs, image super-resolution, and enhancement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge