Yuan Hui
UIERL: Internal-External Representation Learning Network for Underwater Image Enhancement
Jun 14, 2023



Abstract:Underwater image enhancement (UIE) is a meaningful but challenging task, and many learning-based UIE methods have been proposed in recent years. Although much progress has been made, these methods still exist two issues: (1) There exists a significant region-wise quality difference in a single underwater image due to the underwater imaging process, especially in regions with different scene depths. However, existing methods neglect this internal characteristic of underwater images, resulting in inferior performance; (2) Due to the uniqueness of the acquisition approach, underwater image acquisition tools usually capture multiple images in the same or similar scenes. Thus, the underwater images to be enhanced in practical usage are highly correlated. However, when processing a single image, existing methods do not consider the rich external information provided by the related images. There is still room for improvement in their performance. Motivated by these two aspects, we propose a novel internal-external representation learning (UIERL) network to better perform UIE tasks with internal and external information, simultaneously. In the internal representation learning stage, a new depth-based region feature guidance network is designed, including a region segmentation based on scene depth to sense regions with different quality levels, followed by a region-wise space encoder module. With performing region-wise feature learning for regions with different quality separately, the network provides an effective guidance for global features and thus guides intra-image differentiated enhancement. In the external representation learning stage, we first propose an external information extraction network to mine the rich external information in the related images. Then, internal and external features interact with each other via the proposed external-assist-internal module and internal-assist-e
Universal Segmentation of 33 Anatomies
Mar 04, 2022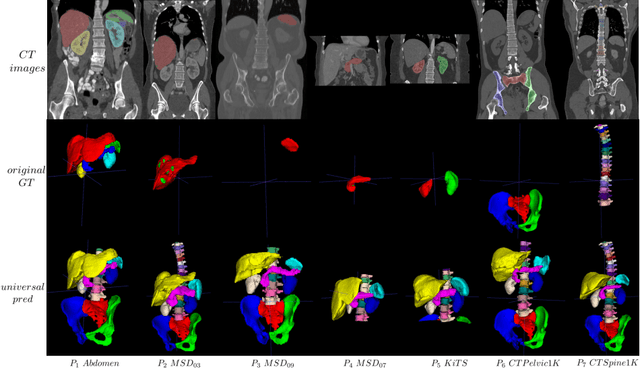
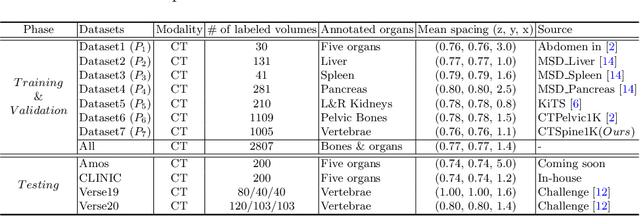
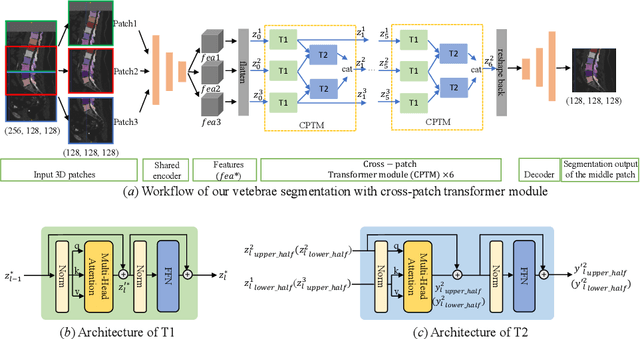
Abstract:In the paper, we present an approach for learning a single model that universally segments 33 anatomical structures, including vertebrae, pelvic bones, and abdominal organs. Our model building has to address the following challenges. Firstly, while it is ideal to learn such a model from a large-scale, fully-annotated dataset, it is practically hard to curate such a dataset. Thus, we resort to learn from a union of multiple datasets, with each dataset containing the images that are partially labeled. Secondly, along the line of partial labelling, we contribute an open-source, large-scale vertebra segmentation dataset for the benefit of spine analysis community, CTSpine1K, boasting over 1,000 3D volumes and over 11K annotated vertebrae. Thirdly, in a 3D medical image segmentation task, due to the limitation of GPU memory, we always train a model using cropped patches as inputs instead a whole 3D volume, which limits the amount of contextual information to be learned. To this, we propose a cross-patch transformer module to fuse more information in adjacent patches, which enlarges the aggregated receptive field for improved segmentation performance. This is especially important for segmenting, say, the elongated spine. Based on 7 partially labeled datasets that collectively contain about 2,800 3D volumes, we successfully learn such a universal model. Finally, we evaluate the universal model on multiple open-source datasets, proving that our model has a good generalization performance and can potentially serve as a solid foundation for downstream tasks.
DuDoTrans: Dual-Domain Transformer Provides More Attention for Sinogram Restoration in Sparse-View CT Reconstruction
Nov 25, 2021



Abstract:While Computed Tomography (CT) reconstruction from X-ray sinograms is necessary for clinical diagnosis, iodine radiation in the imaging process induces irreversible injury, thereby driving researchers to study sparse-view CT reconstruction, that is, recovering a high-quality CT image from a sparse set of sinogram views. Iterative models are proposed to alleviate the appeared artifacts in sparse-view CT images, but the computation cost is too expensive. Then deep-learning-based methods have gained prevalence due to the excellent performances and lower computation. However, these methods ignore the mismatch between the CNN's \textbf{local} feature extraction capability and the sinogram's \textbf{global} characteristics. To overcome the problem, we propose \textbf{Du}al-\textbf{Do}main \textbf{Trans}former (\textbf{DuDoTrans}) to simultaneously restore informative sinograms via the long-range dependency modeling capability of Transformer and reconstruct CT image with both the enhanced and raw sinograms. With such a novel design, reconstruction performance on the NIH-AAPM dataset and COVID-19 dataset experimentally confirms the effectiveness and generalizability of DuDoTrans with fewer involved parameters. Extensive experiments also demonstrate its robustness with different noise-level scenarios for sparse-view CT reconstruction. The code and models are publicly available at https://github.com/DuDoTrans/CODE
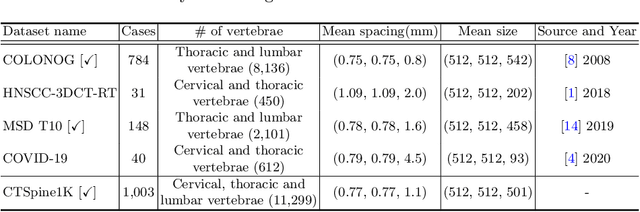
CTSpine1K: A Large-Scale Dataset for Spinal Vertebrae Segmentation in Computed Tomography
Jun 10, 2021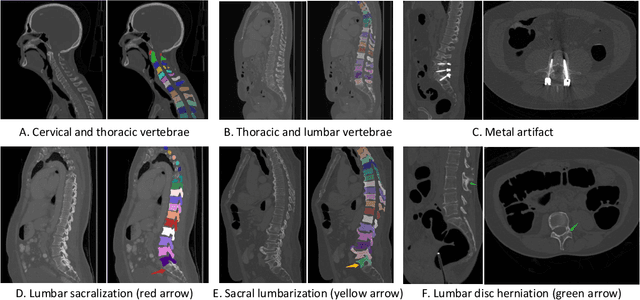
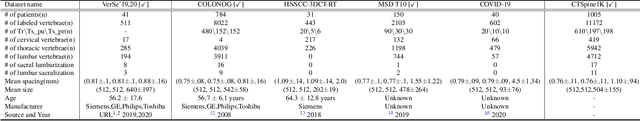
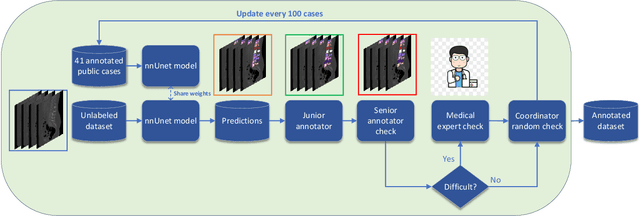
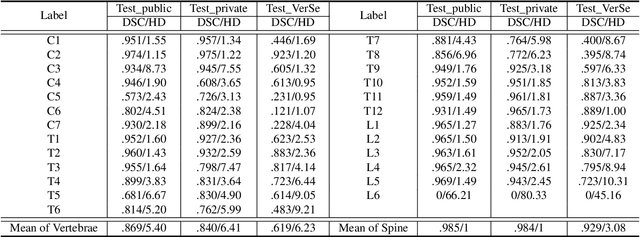
Abstract:Spine-related diseases have high morbidity and cause a huge burden of social cost. Spine imaging is an essential tool for noninvasively visualizing and assessing spinal pathology. Segmenting vertebrae in computed tomography (CT) images is the basis of quantitative medical image analysis for clinical diagnosis and surgery planning of spine diseases. Current publicly available annotated datasets on spinal vertebrae are small in size. Due to the lack of a large-scale annotated spine image dataset, the mainstream deep learning-based segmentation methods, which are data-driven, are heavily restricted. In this paper, we introduce a large-scale spine CT dataset, called CTSpine1K, curated from multiple sources for vertebra segmentation, which contains 1,005 CT volumes with over 11,100 labeled vertebrae belonging to different spinal conditions. Based on this dataset, we conduct several spinal vertebrae segmentation experiments to set the first benchmark. We believe that this large-scale dataset will facilitate further research in many spine-related image analysis tasks, including but not limited to vertebrae segmentation, labeling, 3D spine reconstruction from biplanar radiographs, image super-resolution, and enhancement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge