Mengke Sun
Multi-site Organ Segmentation with Federated Partial Supervision and Site Adaptation
Feb 08, 2023



Abstract:Objective and Impact Statement: Accurate organ segmentation is critical for many clinical applications at different clinical sites, which may have their specific application requirements that concern different organs. Introduction: However, learning high-quality, site-specific organ segmentation models is challenging as it often needs on-site curation of a large number of annotated images. Security concerns further complicate the matter. Methods: The paper aims to tackle these challenges via a two-phase aggregation-then-adaptation approach. The first phase of federated aggregation learns a single multi-organ segmentation model by leveraging the strength of 'bigger data', which are formed by (i) aggregating together datasets from multiple sites that with different organ labels to provide partial supervision, and (ii) conducting partially supervised learning without data breach. The second phase of site adaptation is to transfer the federated multi-organ segmentation model to site-specific organ segmentation models, one model per site, in order to further improve the performance of each site's organ segmentation task. Furthermore, improved marginal loss and exclusion loss functions are used to avoid 'knowledge conflict' problem in a partially supervision mechanism. Results and Conclusion: Extensive experiments on five organ segmentation datasets demonstrate the effectiveness of our multi-site approach, significantly outperforming the site-per-se learned models and achieving the performance comparable to the centrally learned models.
Universal Segmentation of 33 Anatomies
Mar 04, 2022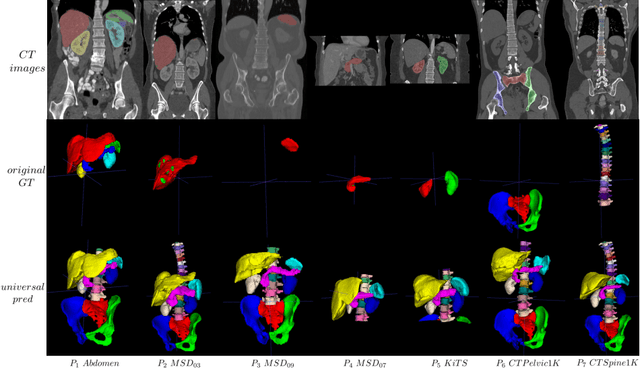
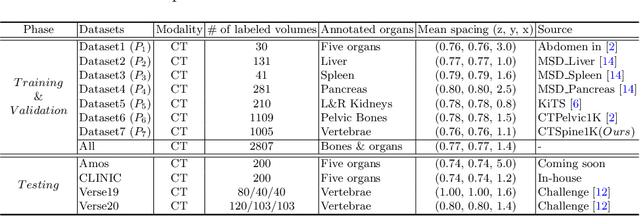
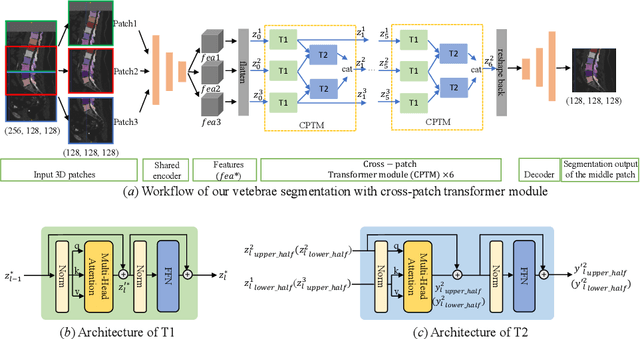
Abstract:In the paper, we present an approach for learning a single model that universally segments 33 anatomical structures, including vertebrae, pelvic bones, and abdominal organs. Our model building has to address the following challenges. Firstly, while it is ideal to learn such a model from a large-scale, fully-annotated dataset, it is practically hard to curate such a dataset. Thus, we resort to learn from a union of multiple datasets, with each dataset containing the images that are partially labeled. Secondly, along the line of partial labelling, we contribute an open-source, large-scale vertebra segmentation dataset for the benefit of spine analysis community, CTSpine1K, boasting over 1,000 3D volumes and over 11K annotated vertebrae. Thirdly, in a 3D medical image segmentation task, due to the limitation of GPU memory, we always train a model using cropped patches as inputs instead a whole 3D volume, which limits the amount of contextual information to be learned. To this, we propose a cross-patch transformer module to fuse more information in adjacent patches, which enlarges the aggregated receptive field for improved segmentation performance. This is especially important for segmenting, say, the elongated spine. Based on 7 partially labeled datasets that collectively contain about 2,800 3D volumes, we successfully learn such a universal model. Finally, we evaluate the universal model on multiple open-source datasets, proving that our model has a good generalization performance and can potentially serve as a solid foundation for downstream tasks.
CTSpine1K: A Large-Scale Dataset for Spinal Vertebrae Segmentation in Computed Tomography
Jun 10, 2021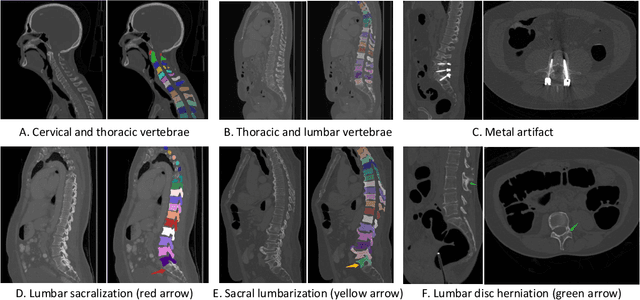
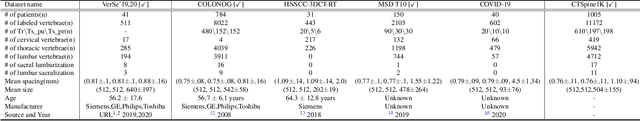
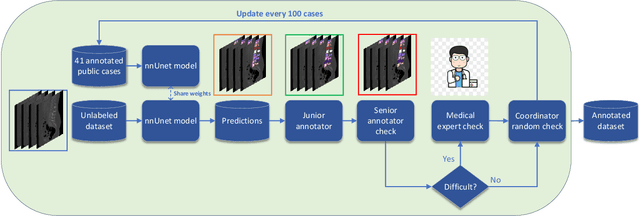
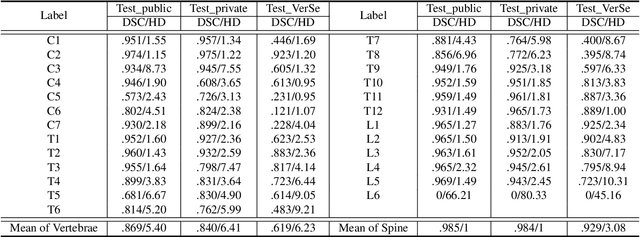
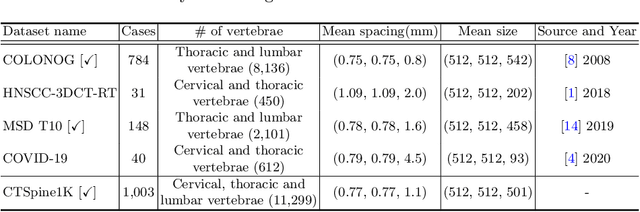
Abstract:Spine-related diseases have high morbidity and cause a huge burden of social cost. Spine imaging is an essential tool for noninvasively visualizing and assessing spinal pathology. Segmenting vertebrae in computed tomography (CT) images is the basis of quantitative medical image analysis for clinical diagnosis and surgery planning of spine diseases. Current publicly available annotated datasets on spinal vertebrae are small in size. Due to the lack of a large-scale annotated spine image dataset, the mainstream deep learning-based segmentation methods, which are data-driven, are heavily restricted. In this paper, we introduce a large-scale spine CT dataset, called CTSpine1K, curated from multiple sources for vertebra segmentation, which contains 1,005 CT volumes with over 11,100 labeled vertebrae belonging to different spinal conditions. Based on this dataset, we conduct several spinal vertebrae segmentation experiments to set the first benchmark. We believe that this large-scale dataset will facilitate further research in many spine-related image analysis tasks, including but not limited to vertebrae segmentation, labeling, 3D spine reconstruction from biplanar radiographs, image super-resolution, and enhancement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge