Xingwang Wu
Learning Incrementally to Segment Multiple Organs in a CT Image
Mar 04, 2022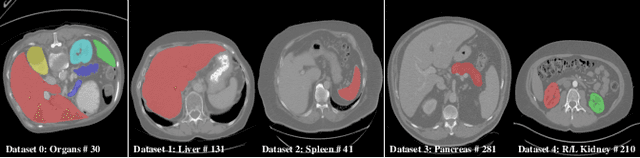
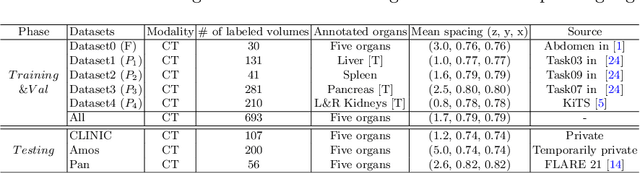
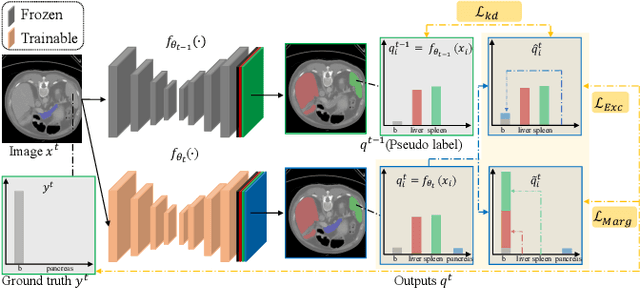
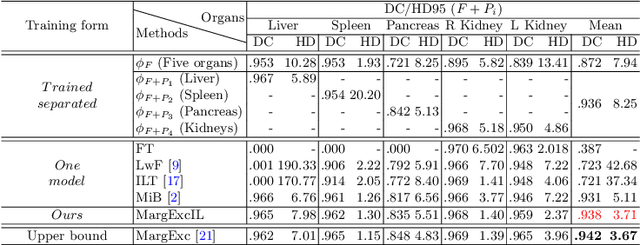
Abstract:There exists a large number of datasets for organ segmentation, which are partially annotated and sequentially constructed. A typical dataset is constructed at a certain time by curating medical images and annotating the organs of interest. In other words, new datasets with annotations of new organ categories are built over time. To unleash the potential behind these partially labeled, sequentially-constructed datasets, we propose to incrementally learn a multi-organ segmentation model. In each incremental learning (IL) stage, we lose the access to previous data and annotations, whose knowledge is assumingly captured by the current model, and gain the access to a new dataset with annotations of new organ categories, from which we learn to update the organ segmentation model to include the new organs. While IL is notorious for its `catastrophic forgetting' weakness in the context of natural image analysis, we experimentally discover that such a weakness mostly disappears for CT multi-organ segmentation. To further stabilize the model performance across the IL stages, we introduce a light memory module and some loss functions to restrain the representation of different categories in feature space, aggregating feature representation of the same class and separating feature representation of different classes. Extensive experiments on five open-sourced datasets are conducted to illustrate the effectiveness of our method.
Radiology Report Generation with a Learned Knowledge Base and Multi-modal Alignment
Dec 30, 2021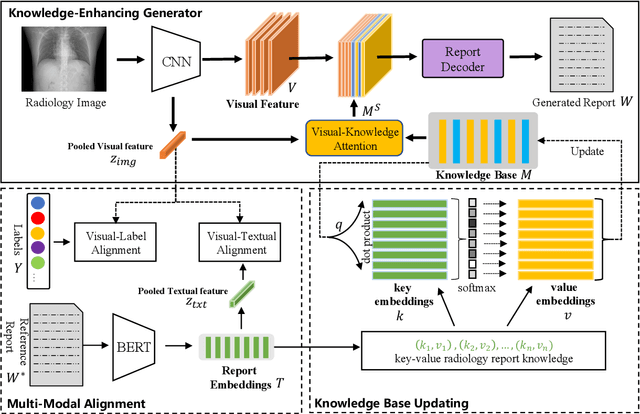
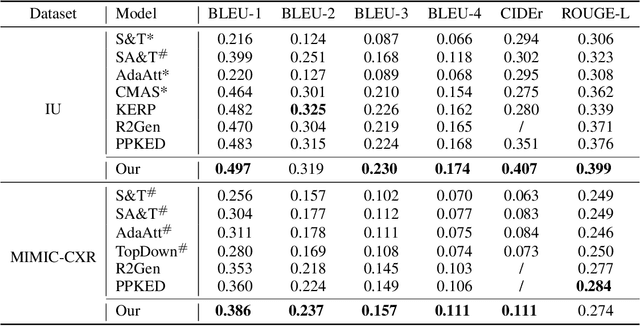
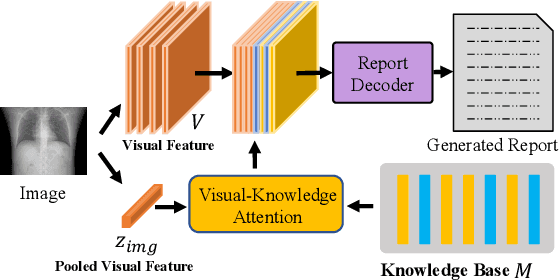
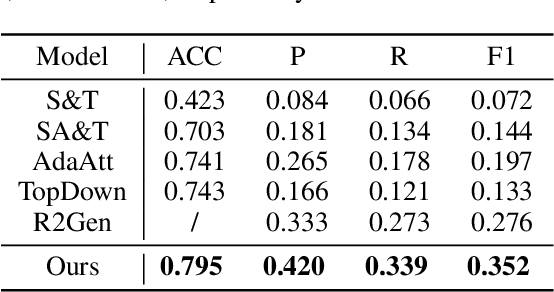
Abstract:In clinics, a radiology report is crucial for guiding a patient's treatment. Unfortunately, report writing imposes a heavy burden on radiologists. To effectively reduce such a burden, we hereby present an automatic, multi-modal approach for report generation from chest x-ray. Our approach, motivated by the observation that the descriptions in radiology reports are highly correlated with the x-ray images, features two distinct modules: (i) Learned knowledge base. To absorb the knowledge embedded in the above-mentioned correlation, we automatically build a knowledge base based on textual embedding. (ii) Multi-modal alignment. To promote the semantic alignment among reports, disease labels and images, we explicitly utilize textual embedding to guide the learning of the visual feature space. We evaluate the performance of the proposed model using metrics from both natural language generation and clinic efficacy on the public IU and MIMIC-CXR datasets. Our ablation study shows that each module contributes to improving the quality of generated reports. Furthermore, with the aid of both modules, our approach clearly outperforms state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge