Ning Wen
Functional Programming Paradigm of Python for Scientific Computation Pipeline Integration
May 27, 2024Abstract:The advent of modern data processing has led to an increasing tendency towards interdisciplinarity, which frequently involves the importation of different technical approaches. Consequently, there is an urgent need for a unified data control system to facilitate the integration of varying libraries. This integration is of profound significance in accelerating prototype verification, optimising algorithm performance and minimising maintenance costs. This paper presents a novel functional programming (FP) paradigm based on the Python architecture and associated suites in programming practice, designed for the integration of pipelines of different data mapping operations. In particular, the solution is intended for the integration of scientific computation flows, which affords a robust yet flexible solution for the aforementioned challenges.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Accurate Prostate Cancer Detection and Segmentation on Biparametric MRI using Non-local Mask R-CNN with Histopathological Ground Truth
Oct 28, 2020



Abstract:Purpose: We aimed to develop deep machine learning (DL) models to improve the detection and segmentation of intraprostatic lesions (IL) on bp-MRI by using whole amount prostatectomy specimen-based delineations. We also aimed to investigate whether transfer learning and self-training would improve results with small amount labelled data. Methods: 158 patients had suspicious lesions delineated on MRI based on bp-MRI, 64 patients had ILs delineated on MRI based on whole mount prostatectomy specimen sections, 40 patients were unlabelled. A non-local Mask R-CNN was proposed to improve the segmentation accuracy. Transfer learning was investigated by fine-tuning a model trained using MRI-based delineations with prostatectomy-based delineations. Two label selection strategies were investigated in self-training. The performance of models was evaluated by 3D detection rate, dice similarity coefficient (DSC), 95 percentile Hausdrauff (95 HD, mm) and true positive ratio (TPR). Results: With prostatectomy-based delineations, the non-local Mask R-CNN with fine-tuning and self-training significantly improved all evaluation metrics. For the model with the highest detection rate and DSC, 80.5% (33/41) of lesions in all Gleason Grade Groups (GGG) were detected with DSC of 0.548[0.165], 95 HD of 5.72[3.17] and TPR of 0.613[0.193]. Among them, 94.7% (18/19) of lesions with GGG > 2 were detected with DSC of 0.604[0.135], 95 HD of 6.26[3.44] and TPR of 0.580[0.190]. Conclusion: DL models can achieve high prostate cancer detection and segmentation accuracy on bp-MRI based on annotations from histologic images. To further improve the performance, more data with annotations of both MRI and whole amount prostatectomy specimens are required.
Improvement of Multiparametric MR Image Segmentation by Augmenting the Data with Generative Adversarial Networks for Glioma Patients
Oct 01, 2019



Abstract:Every year thousands of patients are diagnosed with a glioma, a type of malignant brain tumor. Physicians use MR images as a key tool in the diagnosis and treatment of these patients. Neural networks show great potential to aid physicians in the medical image analysis. This study investigates the use of varying amounts of synthetic brain T1-weighted (T1), post-contrast T1-weighted (T1Gd), T2-weighted (T2), and T2 Fluid Attenuated Inversion Recovery (FLAIR) MR images created by a generative adversarial network to overcome the lack of annotated medical image data in training separate 2D U-Nets to segment enhancing tumor, peritumoral edema, and necrosis (non-enhancing tumor core) regions on gliomas. These synthetic MR images were assessed quantitively (SSIM=0.79) and qualitatively by a physician who found that the synthetic images seem stronger for delineation of structural boundaries but struggle more when gradient is significant, (e.g. edema signal in T2 modalities). Multiple 2D U-Nets were trained with original BraTS data and differing subsets of a quarter, half, three-quarters, and all synthetic MR images. There was not an obvious correlation between the improvement of values of the metrics in separate validation dataset for each structure and amount of synthetic data added, there is a strong correlation between the amount of synthetic data added and the number of best overall validation metrics. In summary, this study showed ability to generate high quality synthetic Flair, T2, T1, and T1CE MR images using the GAN. Using the synthetic MR images showed encouraging results to improve the U-Net segmentation performance which has the potential to address the scarcity of readily available medical images.
A Deep Dive into Understanding Tumor Foci Classification using Multiparametric MRI Based on Convolutional Neural Network
Apr 04, 2019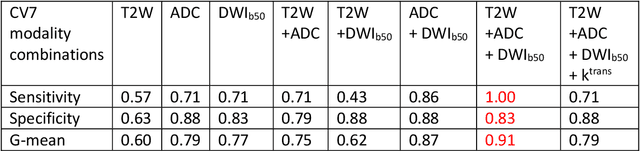
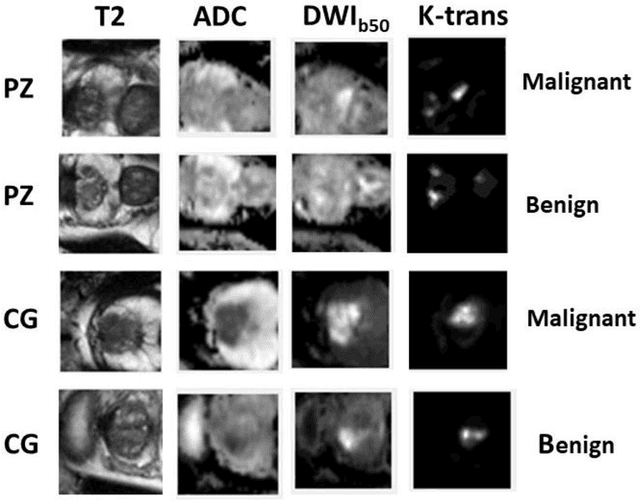
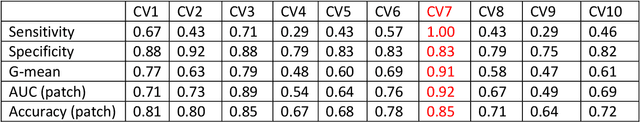
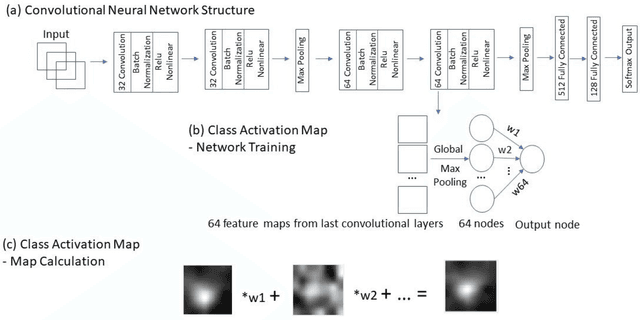
Abstract:Data scarcity has refrained deep learning models from making greater progress in prostate images analysis using multiparametric MRI. In this paper, an efficient convolutional neural network (CNN) was developed to classify lesion malignancy for prostate cancer patients, based on which model interpretation was systematically analyzed to bridge the gap between natural images and MR images, which is the first one of its kind in the literature. The problem of small sample size was addressed and successfully tackled by feeding the intermediate features into a traditional classification algorithm known as weighted extreme learning machine, with imbalanced distribution among output categories taken into consideration. Model trained on public data set was able to generalize to data from an independent institution to make accurate prediction. The generated saliency map was found to overlay well with the lesion and could benefit clinicians for diagnosing purpose.
Segmentation of the Prostatic Gland and the Intraprostatic Lesions on Multiparametic MRI Using Mask-RCNN
Apr 04, 2019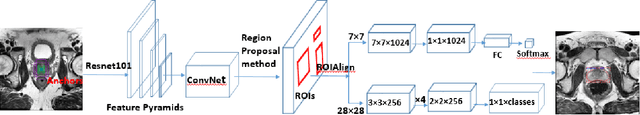
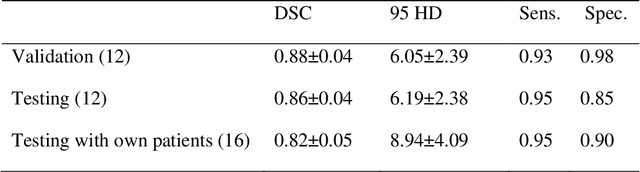
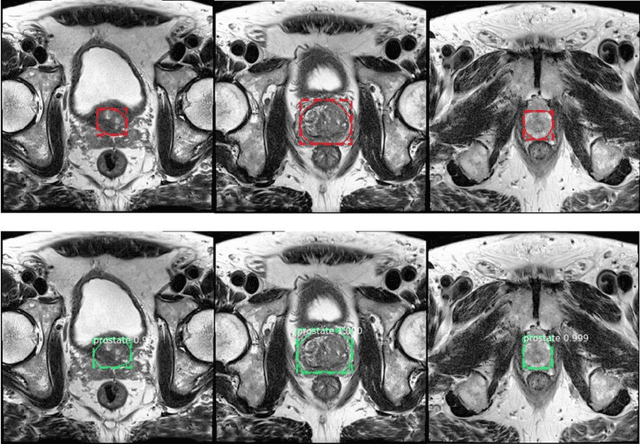
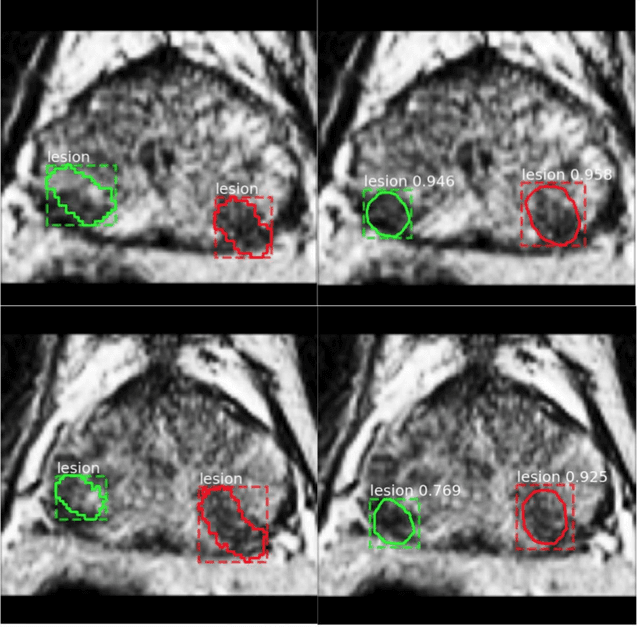
Abstract:Prostate cancer (PCa) is the most common cancer in men in the United States. Multiparametic magnetic resonance imaging (mp-MRI) has been explored by many researchers to targeted prostate biopsies and radiation therapy. However, assessment on mp-MRI can be subjective, development of computer-aided diagnosis systems to automatically delineate the prostate gland and the intraprostratic lesions (ILs) becomes important to facilitate with radiologists in clinical practice. In this paper, we first study the implementation of the Mask-RCNN model to segment the prostate and ILs. We trained and evaluated models on 120 patients from two different cohorts of patients. We also used 2D U-Net and 3D U-Net as benchmarks to segment the prostate and compared the model's performance. The contour variability of ILs using the algorithm was also benchmarked against the interobserver variability between two different radiation oncologists on 19 patients. Our results indicate that the Mask-RCNN model is able to reach state-of-art performance in the prostate segmentation and outperforms several competitive baselines in ILs segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge