Netanell Avisdris
A multi-centre, multi-device benchmark dataset for landmark-based comprehensive fetal biometry
Dec 19, 2025Abstract:Accurate fetal growth assessment from ultrasound (US) relies on precise biometry measured by manually identifying anatomical landmarks in standard planes. Manual landmarking is time-consuming, operator-dependent, and sensitive to variability across scanners and sites, limiting the reproducibility of automated approaches. There is a need for multi-source annotated datasets to develop artificial intelligence-assisted fetal growth assessment methods. To address this bottleneck, we present an open, multi-centre, multi-device benchmark dataset of fetal US images with expert anatomical landmark annotations for clinically used fetal biometric measurements. These measurements include head bi-parietal and occipito-frontal diameters, abdominal transverse and antero-posterior diameters, and femoral length. The dataset comprises 4,513 de-identified US images from 1,904 subjects acquired at three clinical sites using seven different US devices. We provide standardised, subject-disjoint train/test splits, evaluation code, and baseline results to enable fair and reproducible comparison of methods. Using an automatic biometry model, we quantify domain shift and demonstrate that training and evaluation confined to a single centre substantially overestimate performance relative to multi-centre testing. To the best of our knowledge, this is the first publicly available multi-centre, multi-device, landmark-annotated dataset that covers all primary fetal biometry measures, providing a robust benchmark for domain adaptation and multi-centre generalisation in fetal biometry and enabling more reliable AI-assisted fetal growth assessment across centres. All data, annotations, training code, and evaluation pipelines are made publicly available.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Bounded Future MS-TCN++ for surgical gesture recognition
Oct 05, 2022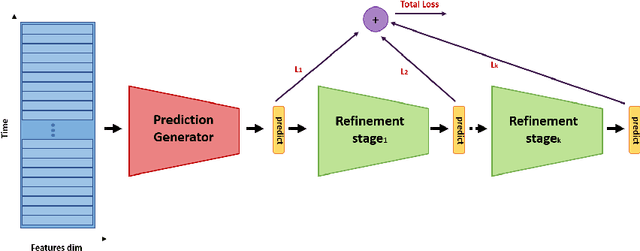

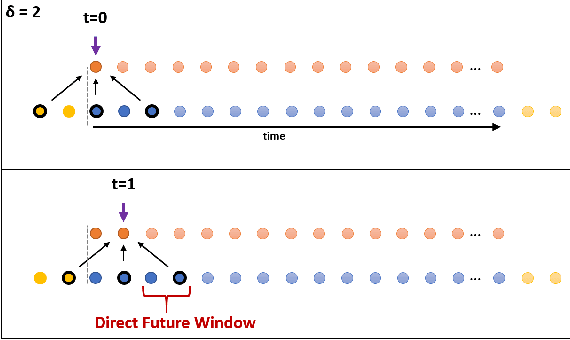
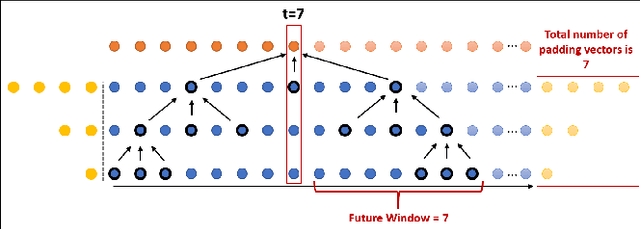
Abstract:In recent times there is a growing development of video based applications for surgical purposes. Part of these applications can work offline after the end of the procedure, other applications must react immediately. However, there are cases where the response should be done during the procedure but some delay is acceptable. In the literature, the online-offline performance gap is known. Our goal in this study was to learn the performance-delay trade-off and design an MS-TCN++-based algorithm that can utilize this trade-off. To this aim, we used our open surgery simulation data-set containing 96 videos of 24 participants that perform a suturing task on a variable tissue simulator. In this study, we used video data captured from the side view. The Networks were trained to identify the performed surgical gestures. The naive approach is to reduce the MS-TCN++ depth, as a result, the receptive field is reduced, and also the number of required future frames is also reduced. We showed that this method is sub-optimal, mainly in the small delay cases. The second method was to limit the accessible future in each temporal convolution. This way, we have flexibility in the network design and as a result, we achieve significantly better performance than in the naive approach.
Automatic fetal fat quantification from MRI
Sep 08, 2022



Abstract:Normal fetal adipose tissue (AT) development is essential for perinatal well-being. AT, or simply fat, stores energy in the form of lipids. Malnourishment may result in excessive or depleted adiposity. Although previous studies showed a correlation between the amount of AT and perinatal outcome, prenatal assessment of AT is limited by lacking quantitative methods. Using magnetic resonance imaging (MRI), 3D fat- and water-only images of the entire fetus can be obtained from two point Dixon images to enable AT lipid quantification. This paper is the first to present a methodology for developing a deep learning based method for fetal fat segmentation based on Dixon MRI. It optimizes radiologists' manual fetal fat delineation time to produce annotated training dataset. It consists of two steps: 1) model-based semi-automatic fetal fat segmentations, reviewed and corrected by a radiologist; 2) automatic fetal fat segmentation using DL networks trained on the resulting annotated dataset. Three DL networks were trained. We show a significant improvement in segmentation times (3:38 hours to < 1 hour) and observer variability (Dice of 0.738 to 0.906) compared to manual segmentation. Automatic segmentation of 24 test cases with the 3D Residual U-Net, nn-UNet and SWIN-UNetR transformer networks yields a mean Dice score of 0.863, 0.787 and 0.856, respectively. These results are better than the manual observer variability, and comparable to automatic adult and pediatric fat segmentation. A radiologist reviewed and corrected six new independent cases segmented using the best performing network, resulting in a Dice score of 0.961 and a significantly reduced correction time of 15:20 minutes. Using these novel segmentation methods and short MRI acquisition time, whole body subcutaneous lipids can be quantified for individual fetuses in the clinic and large-cohort research.
BiometryNet: Landmark-based Fetal Biometry Estimation from Standard Ultrasound Planes
Jun 29, 2022



Abstract:Fetal growth assessment from ultrasound is based on a few biometric measurements that are performed manually and assessed relative to the expected gestational age. Reliable biometry estimation depends on the precise detection of landmarks in standard ultrasound planes. Manual annotation can be time-consuming and operator dependent task, and may results in high measurements variability. Existing methods for automatic fetal biometry rely on initial automatic fetal structure segmentation followed by geometric landmark detection. However, segmentation annotations are time-consuming and may be inaccurate, and landmark detection requires developing measurement-specific geometric methods. This paper describes BiometryNet, an end-to-end landmark regression framework for fetal biometry estimation that overcomes these limitations. It includes a novel Dynamic Orientation Determination (DOD) method for enforcing measurement-specific orientation consistency during network training. DOD reduces variabilities in network training, increases landmark localization accuracy, thus yields accurate and robust biometric measurements. To validate our method, we assembled a dataset of 3,398 ultrasound images from 1,829 subjects acquired in three clinical sites with seven different ultrasound devices. Comparison and cross-validation of three different biometric measurements on two independent datasets shows that BiometryNet is robust and yields accurate measurements whose errors are lower than the clinically permissible errors, outperforming other existing automated biometry estimation methods. Code is available at https://github.com/netanellavisdris/fetalbiometry.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022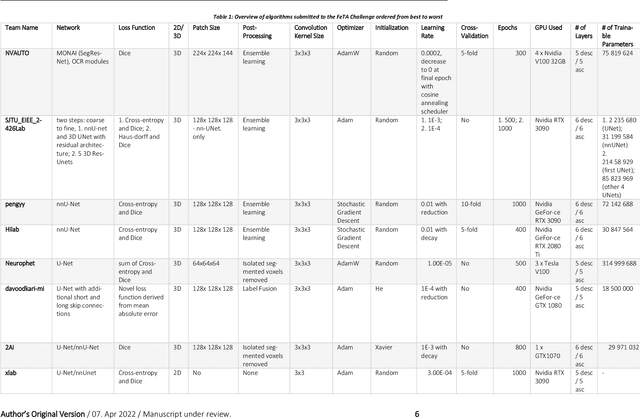
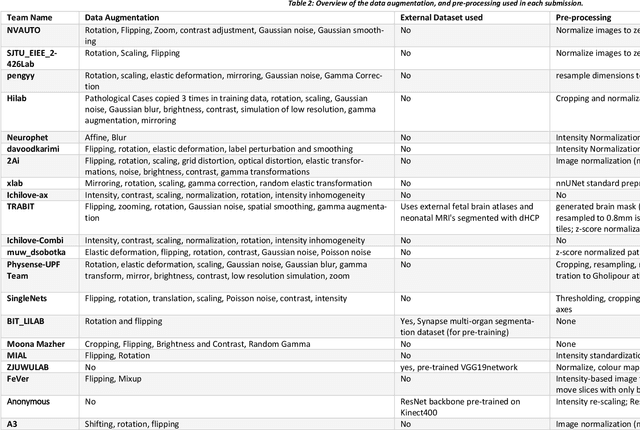
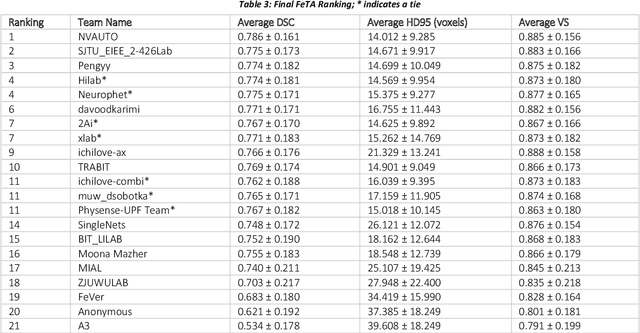
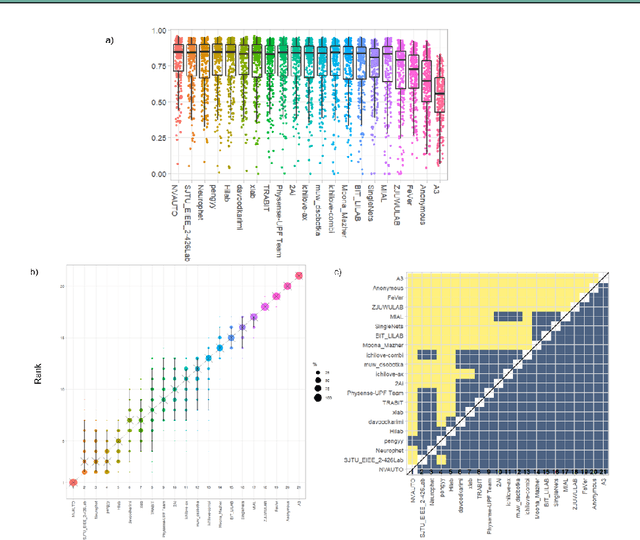
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
Automatic linear measurements of the fetal brain on MRI with deep neural networks
Jun 15, 2021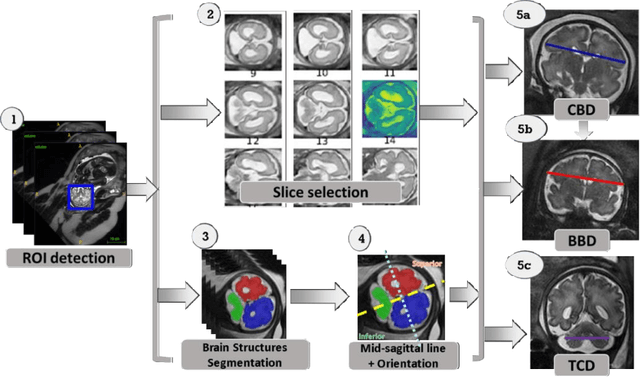
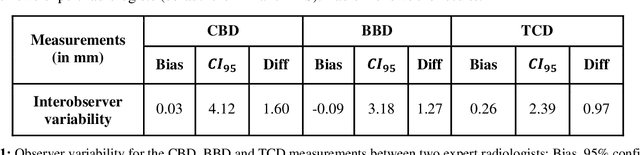
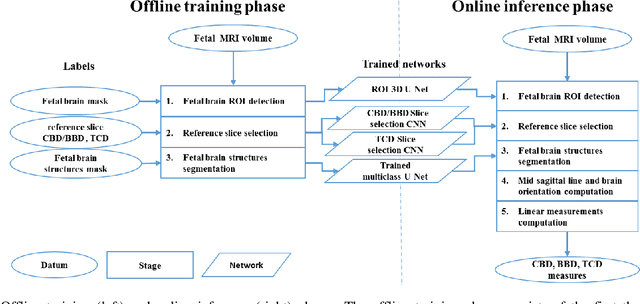
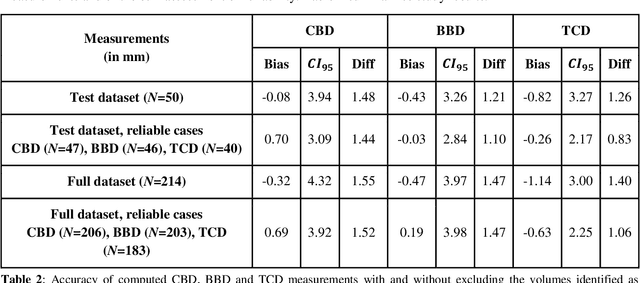
Abstract:Timely, accurate and reliable assessment of fetal brain development is essential to reduce short and long-term risks to fetus and mother. Fetal MRI is increasingly used for fetal brain assessment. Three key biometric linear measurements important for fetal brain evaluation are Cerebral Biparietal Diameter (CBD), Bone Biparietal Diameter (BBD), and Trans-Cerebellum Diameter (TCD), obtained manually by expert radiologists on reference slices, which is time consuming and prone to human error. The aim of this study was to develop a fully automatic method computing the CBD, BBD and TCD measurements from fetal brain MRI. The input is fetal brain MRI volumes which may include the fetal body and the mother's abdomen. The outputs are the measurement values and reference slices on which the measurements were computed. The method, which follows the manual measurements principle, consists of five stages: 1) computation of a Region Of Interest that includes the fetal brain with an anisotropic 3D U-Net classifier; 2) reference slice selection with a Convolutional Neural Network; 3) slice-wise fetal brain structures segmentation with a multiclass U-Net classifier; 4) computation of the fetal brain midsagittal line and fetal brain orientation, and; 5) computation of the measurements. Experimental results on 214 volumes for CBD, BBD and TCD measurements yielded a mean $L_1$ difference of 1.55mm, 1.45mm and 1.23mm respectively, and a Bland-Altman 95% confidence interval ($CI_{95}$) of 3.92mm, 3.98mm and 2.25mm respectively. These results are similar to the manual inter-observer variability. The proposed automatic method for computing biometric linear measurements of the fetal brain from MR imaging achieves human level performance. It has the potential of being a useful method for the assessment of fetal brain biometry in normal and pathological cases, and of improving routine clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge