Mathieu Andreux
FedECA: A Federated External Control Arm Method for Causal Inference with Time-To-Event Data in Distributed Settings
Nov 28, 2023Abstract:External control arms (ECA) can inform the early clinical development of experimental drugs and provide efficacy evidence for regulatory approval in non-randomized settings. However, the main challenge of implementing ECA lies in accessing real-world data or historical clinical trials. Indeed, data sharing is often not feasible due to privacy considerations related to data leaving the original collection centers, along with pharmaceutical companies' competitive motives. In this paper, we leverage a privacy-enhancing technology called federated learning (FL) to remove some of the barriers to data sharing. We introduce a federated learning inverse probability of treatment weighted (IPTW) method for time-to-event outcomes called FedECA which eases the implementation of ECA by limiting patients' data exposure. We show with extensive experiments that FedECA outperforms its closest competitor, matching-adjusted indirect comparison (MAIC), in terms of statistical power and ability to balance the treatment and control groups. To encourage the use of such methods, we publicly release our code which relies on Substra, an open-source FL software with proven experience in privacy-sensitive contexts.
FLamby: Datasets and Benchmarks for Cross-Silo Federated Learning in Realistic Healthcare Settings
Oct 10, 2022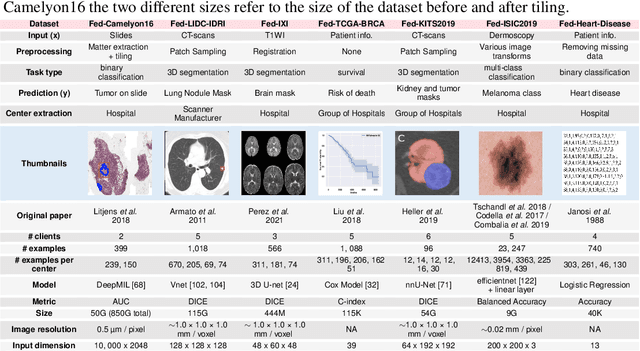
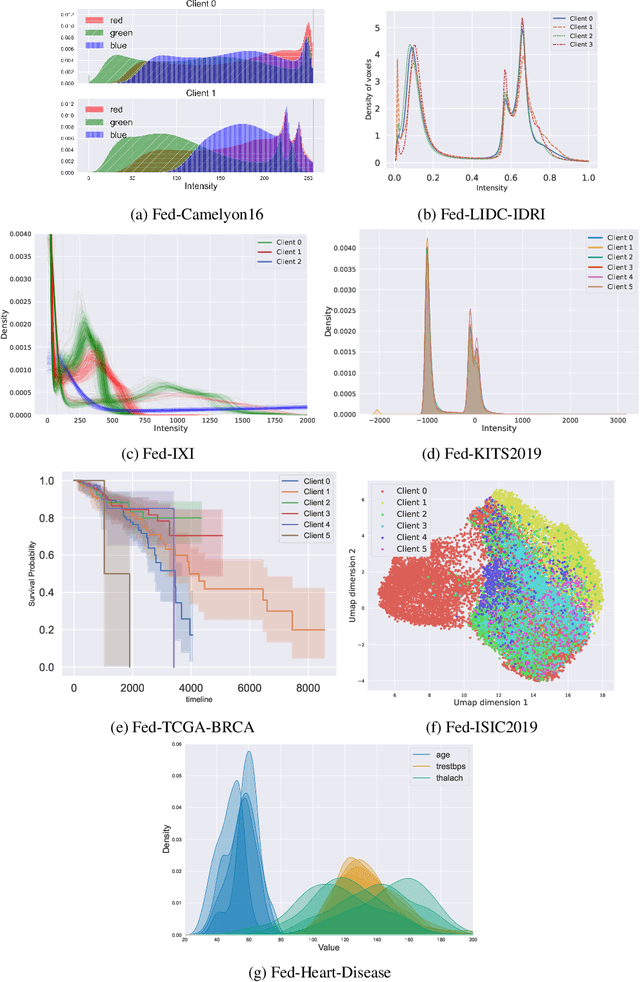
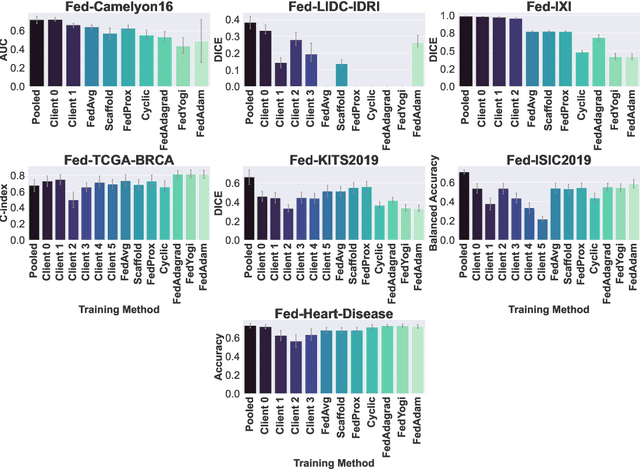
Abstract:Federated Learning (FL) is a novel approach enabling several clients holding sensitive data to collaboratively train machine learning models, without centralizing data. The cross-silo FL setting corresponds to the case of few ($2$--$50$) reliable clients, each holding medium to large datasets, and is typically found in applications such as healthcare, finance, or industry. While previous works have proposed representative datasets for cross-device FL, few realistic healthcare cross-silo FL datasets exist, thereby slowing algorithmic research in this critical application. In this work, we propose a novel cross-silo dataset suite focused on healthcare, FLamby (Federated Learning AMple Benchmark of Your cross-silo strategies), to bridge the gap between theory and practice of cross-silo FL. FLamby encompasses 7 healthcare datasets with natural splits, covering multiple tasks, modalities, and data volumes, each accompanied with baseline training code. As an illustration, we additionally benchmark standard FL algorithms on all datasets. Our flexible and modular suite allows researchers to easily download datasets, reproduce results and re-use the different components for their research. FLamby is available at~\url{www.github.com/owkin/flamby}.
SecureFedYJ: a safe feature Gaussianization protocol for Federated Learning
Oct 04, 2022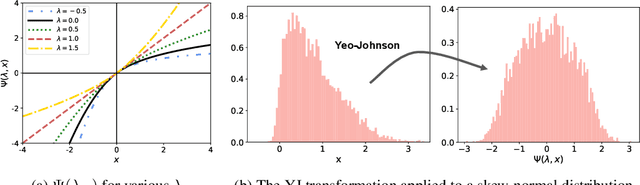
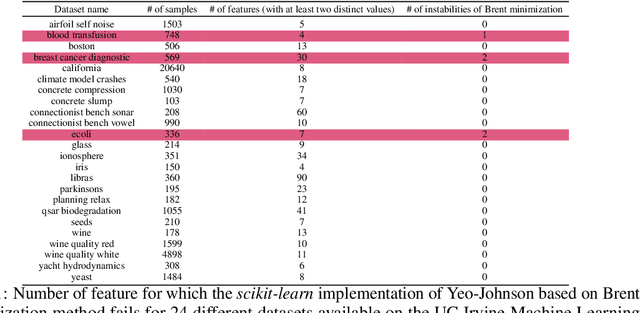
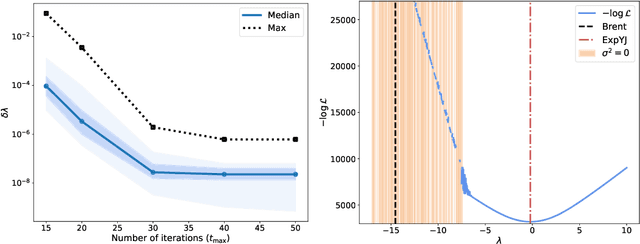
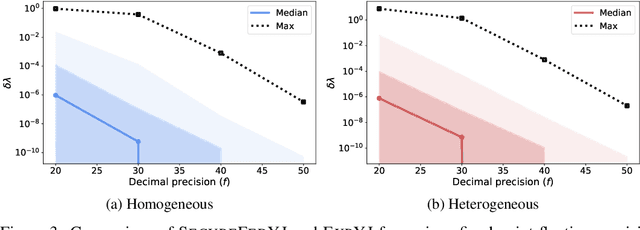
Abstract:The Yeo-Johnson (YJ) transformation is a standard parametrized per-feature unidimensional transformation often used to Gaussianize features in machine learning. In this paper, we investigate the problem of applying the YJ transformation in a cross-silo Federated Learning setting under privacy constraints. For the first time, we prove that the YJ negative log-likelihood is in fact convex, which allows us to optimize it with exponential search. We numerically show that the resulting algorithm is more stable than the state-of-the-art approach based on the Brent minimization method. Building on this simple algorithm and Secure Multiparty Computation routines, we propose SecureFedYJ, a federated algorithm that performs a pooled-equivalent YJ transformation without leaking more information than the final fitted parameters do. Quantitative experiments on real data demonstrate that, in addition to being secure, our approach reliably normalizes features across silos as well as if data were pooled, making it a viable approach for safe federated feature Gaussianization.
Differentially Private Federated Learning for Cancer Prediction
Jan 08, 2021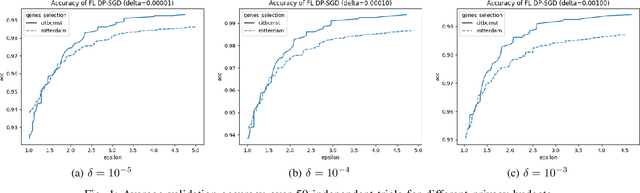
Abstract:Since 2014, the NIH funded iDASH (integrating Data for Analysis, Anonymization, SHaring) National Center for Biomedical Computing has hosted yearly competitions on the topic of private computing for genomic data. For one track of the 2020 iteration of this competition, participants were challenged to produce an approach to federated learning (FL) training of genomic cancer prediction models using differential privacy (DP), with submissions ranked according to held-out test accuracy for a given set of DP budgets. More precisely, in this track, we are tasked with training a supervised model for the prediction of breast cancer occurrence from genomic data split between two virtual centers while ensuring data privacy with respect to model transfer via DP. In this article, we present our 3rd place submission to this competition. During the competition, we encountered two main challenges discussed in this article: i) ensuring correctness of the privacy budget evaluation and ii) achieving an acceptable trade-off between prediction performance and privacy budget.
Siloed Federated Learning for Multi-Centric Histopathology Datasets
Aug 17, 2020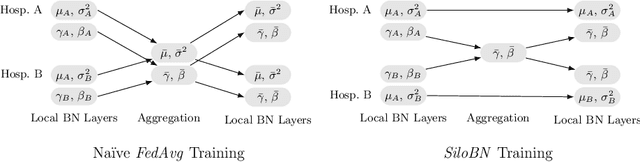
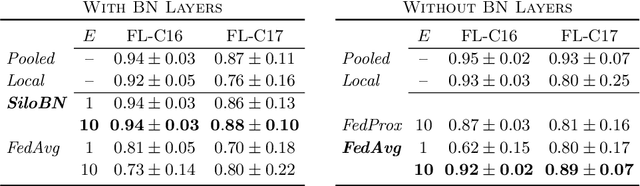
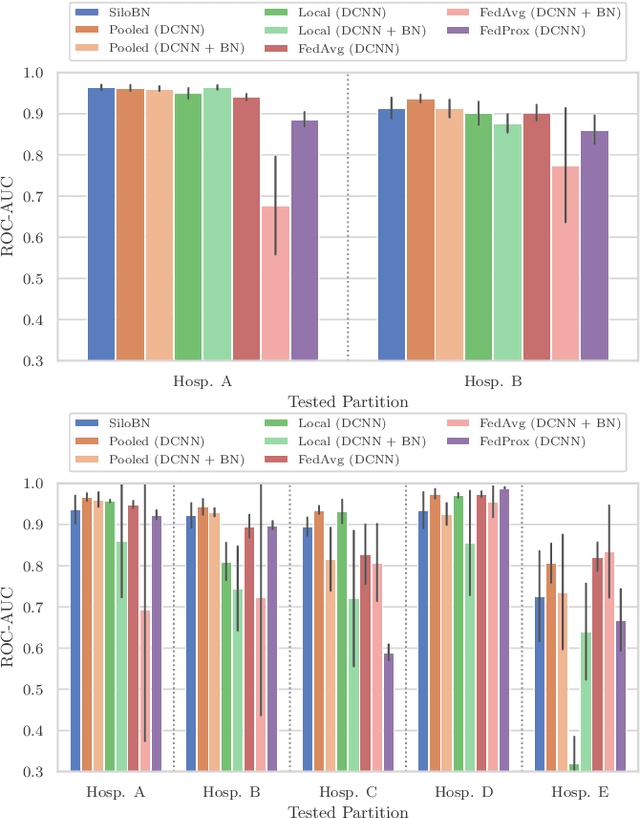

Abstract:While federated learning is a promising approach for training deep learning models over distributed sensitive datasets, it presents new challenges for machine learning, especially when applied in the medical domain where multi-centric data heterogeneity is common. Building on previous domain adaptation works, this paper proposes a novel federated learning approach for deep learning architectures via the introduction of local-statistic batch normalization (BN) layers, resulting in collaboratively-trained, yet center-specific models. This strategy improves robustness to data heterogeneity while also reducing the potential for information leaks by not sharing the center-specific layer activation statistics. We benchmark the proposed method on the classification of tumorous histopathology image patches extracted from the Camelyon16 and Camelyon17 datasets. We show that our approach compares favorably to previous state-of-the-art methods, especially for transfer learning across datasets.
Federated Survival Analysis with Discrete-Time Cox Models
Jun 16, 2020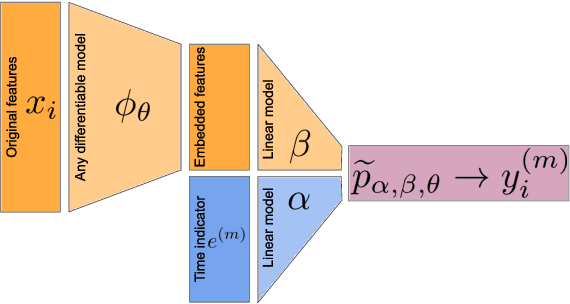
Abstract:Building machine learning models from decentralized datasets located in different centers with federated learning (FL) is a promising approach to circumvent local data scarcity while preserving privacy. However, the prominent Cox proportional hazards (PH) model, used for survival analysis, does not fit the FL framework, as its loss function is non-separable with respect to the samples. The na\"ive method to bypass this non-separability consists in calculating the losses per center, and minimizing their sum as an approximation of the true loss. We show that the resulting model may suffer from important performance loss in some adverse settings. Instead, we leverage the discrete-time extension of the Cox PH model to formulate survival analysis as a classification problem with a separable loss function. Using this approach, we train survival models using standard FL techniques on synthetic data, as well as real-world datasets from The Cancer Genome Atlas (TCGA), showing similar performance to a Cox PH model trained on aggregated data. Compared to previous works, the proposed method is more communication-efficient, more generic, and more amenable to using privacy-preserving techniques.
Kymatio: Scattering Transforms in Python
Dec 28, 2018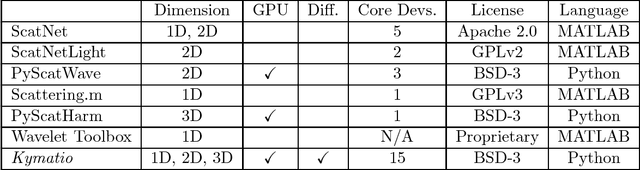
Abstract:The wavelet scattering transform is an invariant signal representation suitable for many signal processing and machine learning applications. We present the Kymatio software package, an easy-to-use, high-performance Python implementation of the scattering transform in 1D, 2D, and 3D that is compatible with modern deep learning frameworks. All transforms may be executed on a GPU (in addition to CPU), offering a considerable speed up over CPU implementations. The package also has a small memory footprint, resulting inefficient memory usage. The source code, documentation, and examples are available undera BSD license at https://www.kymat.io/
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge