Joakim Andén
Moment-based Posterior Sampling for Multi-reference Alignment
Oct 14, 2025


Abstract:We propose a Bayesian approach to the problem of multi-reference alignment -- the recovery of signals from noisy, randomly shifted observations. While existing frequentist methods accurately recover the signal at arbitrarily low signal-to-noise ratios, they require a large number of samples to do so. In contrast, our proposed method leverages diffusion models as data-driven plug-and-play priors, conditioning these on the sample power spectrum (a shift-invariant statistic) enabling both accurate posterior sampling and uncertainty quantification. The use of an appropriate prior significantly reduces the required number of samples, as illustrated in simulation experiments with comparisons to state-of-the-art methods such as expectation--maximization and bispectrum inversion. These findings establish our approach as a promising framework for other orbit recovery problems, such as cryogenic electron microscopy (cryo-EM).
Joint Denoising of Cryo-EM Projection Images using Polar Transformers
Jun 12, 2025Abstract:Deep neural networks~(DNNs) have proven powerful for denoising, but they are ultimately of limited use in high-noise settings, such as for cryogenic electron microscopy~(cryo-EM) projection images. In this setting, however, datasets contain a large number of projections of the same molecule, each taken from a different viewing direction. This redundancy of information is useful in traditional denoising techniques known as class averaging methods, where images are clustered, aligned, and then averaged to reduce the noise level. We present a neural network architecture based on transformers that extends these class averaging methods by simultaneously clustering, aligning, and denoising cryo-EM images. Results on synthetic data show accurate denoising performance using this architecture, reducing the relative mean squared error (MSE) single-image DNNs by $45\%$ at a signal-to-noise (SNR) of $0.03$.
A continuous boostlet transform for acoustic waves in space-time
Mar 17, 2024



Abstract:Sparse representation systems that encode the signal architecture have had an exceptional impact on sampling and compression paradigms. Remarkable examples are multi-scale directional systems, which, similar to our vision system, encode the underlying architecture of natural images with sparse features. Inspired by this philosophy, the present study introduces a representation system for acoustic waves in 2D space-time, referred to as the boostlet transform, which encodes sparse features of natural acoustic fields with the Poincar\'e group and isotropic dilations. Continuous boostlets, $\psi_{a,\theta,\tau}(\varsigma) = a^{-1} \psi \left(D_a^{-1} B_\theta^{-1}(\varsigma-\tau)\right) \in L^2(\mathbb{R}^2)$, are spatiotemporal functions parametrized with dilations $a > 0$, Lorentz boosts $\theta \in \mathbb{R}$, and translations $\smash{\tau \in \mathbb{R}^2}$ in space--time. The admissibility condition requires that boostlets are supported away from the acoustic radiation cone, i.e., have phase velocities other than the speed of sound, resulting in a peculiar scaling function. The continuous boostlet transform is an isometry for $L^2(\mathbb{R}^2)$, and a sparsity analysis with experimentally measured fields indicates that boostlet coefficients decay faster than wavelets, curvelets, wave atoms, and shearlets. The uncertainty principles and minimizers associated with the boostlet transform are derived and interpreted physically.
Mean-Field Microcanonical Gradient Descent
Mar 13, 2024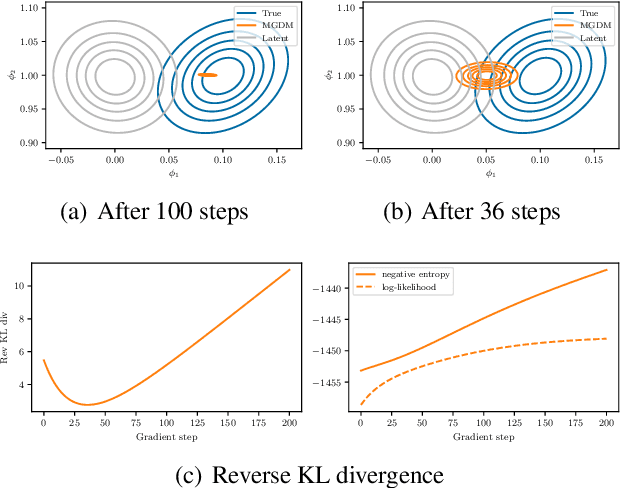
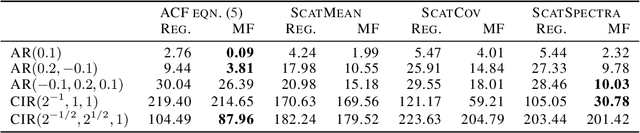
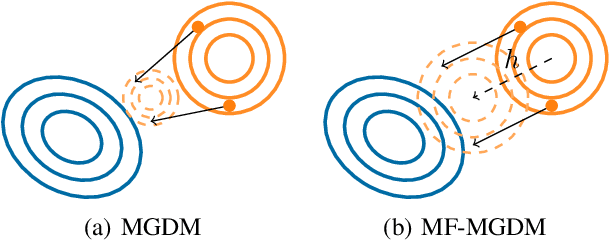
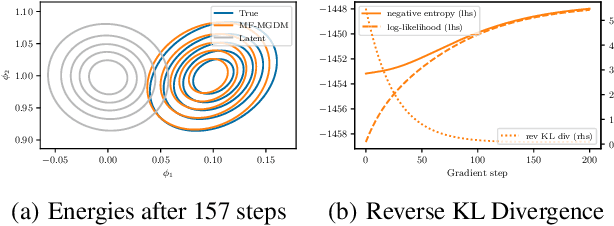
Abstract:Microcanonical gradient descent is a sampling procedure for energy-based models allowing for efficient sampling of distributions in high dimension. It works by transporting samples from a high-entropy distribution, such as Gaussian white noise, to a low-energy region using gradient descent. We put this model in the framework of normalizing flows, showing how it can often overfit by losing an unnecessary amount of entropy in the descent. As a remedy, we propose a mean-field microcanonical gradient descent that samples several weakly coupled data points simultaneously, allowing for better control of the entropy loss while paying little in terms of likelihood fit. We study these models in the context of financial time series, illustrating the improvements on both synthetic and real data.
cuFINUFFT: a load-balanced GPU library for general-purpose nonuniform FFTs
Feb 16, 2021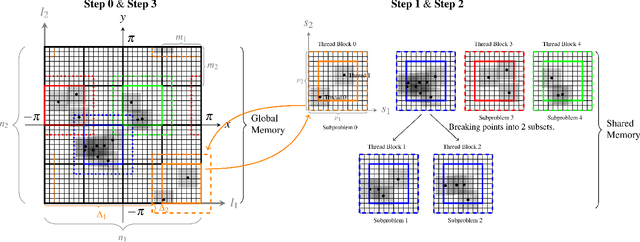

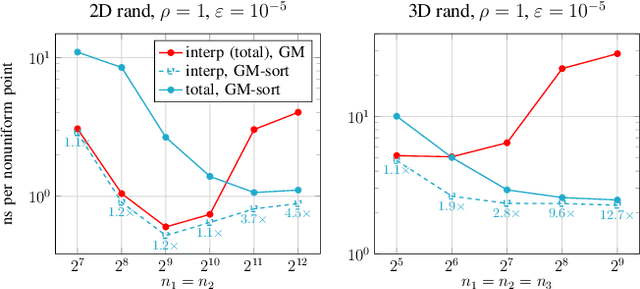
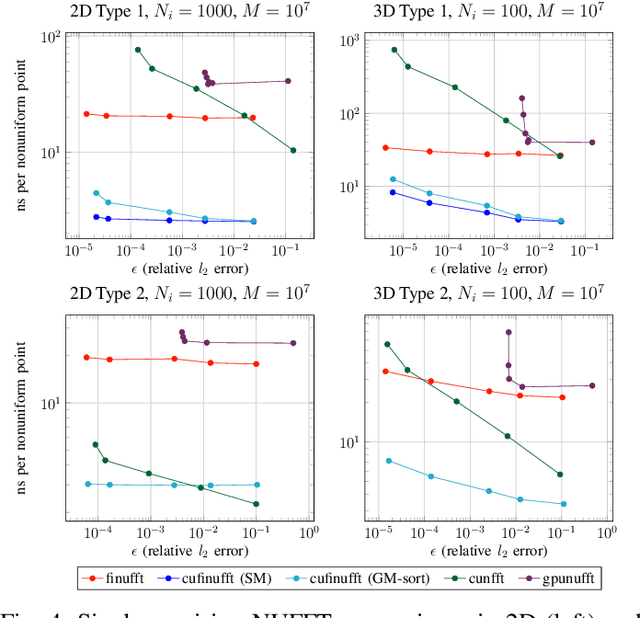
Abstract:Nonuniform fast Fourier transforms dominate the computational cost in many applications including image reconstruction and signal processing. We thus present a general-purpose GPU-based CUDA library for type 1 (nonuniform to uniform) and type 2 (uniform to nonuniform) transforms in dimensions 2 and 3, in single or double precision. It achieves high performance for a given user-requested accuracy, regardless of the distribution of nonuniform points, via cache-aware point reordering, and load-balanced blocked spreading in shared memory. At low accuracies, this gives on-GPU throughputs around $10^9$ nonuniform points per second, and (even including host-device transfer) is typically 4-10$\times$ faster than the latest parallel CPU code FINUFFT (at 28 threads). It is competitive with two established GPU codes, being up to 90$\times$ faster at high accuracy and/or type 1 clustered point distributions. Finally we demonstrate a 6-18$\times$ speedup versus CPU in an X-ray diffraction 3D iterative reconstruction task at $10^{-12}$ accuracy, observing excellent multi-GPU weak scaling up to one rank per GPU.
Time-Frequency Scattering Accurately Models Auditory Similarities Between Instrumental Playing Techniques
Jul 21, 2020



Abstract:Instrumental playing techniques such as vibratos, glissandos, and trills often denote musical expressivity, both in classical and folk contexts. However, most existing approaches to music similarity retrieval fail to describe timbre beyond the so-called ``ordinary'' technique, use instrument identity as a proxy for timbre quality, and do not allow for customization to the perceptual idiosyncrasies of a new subject. In this article, we ask 31 human subjects to organize 78 isolated notes into a set of timbre clusters. Analyzing their responses suggests that timbre perception operates within a more flexible taxonomy than those provided by instruments or playing techniques alone. In addition, we propose a machine listening model to recover the cluster graph of auditory similarities across instruments, mutes, and techniques. Our model relies on joint time--frequency scattering features to extract spectrotemporal modulations as acoustic features. Furthermore, it minimizes triplet loss in the cluster graph by means of the large-margin nearest neighbor (LMNN) metric learning algorithm. Over a dataset of 9346 isolated notes, we report a state-of-the-art average precision at rank five (AP@5) of $99.0\%\pm1$. An ablation study demonstrates that removing either the joint time--frequency scattering transform or the metric learning algorithm noticeably degrades performance.
Bias and variance reduction and denoising for CTF Estimation
Aug 09, 2019

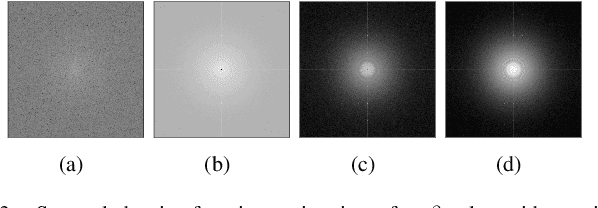

Abstract:When using an electron microscope for imaging of particles embedded in vitreous ice, the objective lens will inevitably corrupt the projection images. This corruption manifests as a band-pass filter on the micrograph. In addition, it causes the phase of several frequency bands to be flipped and distorts frequency bands. As a precursor to compensating for this distortion, the corrupting point spread function, which is termed the contrast transfer function (CTF) in reciprocal space, must be estimated. In this paper, we will present a novel method for CTF estimation. Our method is based on the multi-taper method for power spectral density estimation, which aims to reduce the bias and variance of the estimator. Furthermore, we use known properties of the CTF and of the background of the power spectrum to increase the accuracy of our estimation. We will show that the resulting estimates capture the zero-crossings of the CTF in the low-mid frequency range.
Hyper-Molecules: on the Representation and Recovery of Dynamical Structures, with Application to Flexible Macro-Molecular Structures in Cryo-EM
Jul 02, 2019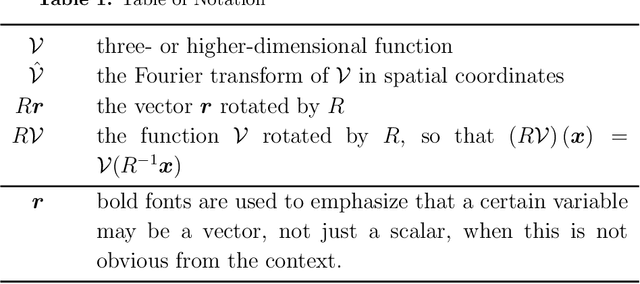
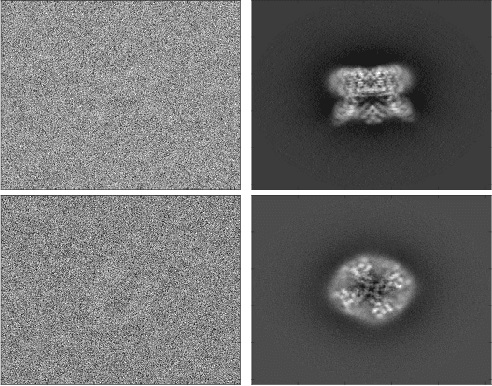

Abstract:Cryo-electron microscopy (cryo-EM), the subject of the 2017 Nobel Prize in Chemistry, is a technology for determining the 3-D structure of macromolecules from many noisy 2-D projections of instances of these macromolecules, whose orientations and positions are unknown. The molecular structures are not rigid objects, but flexible objects involved in dynamical processes. The different conformations are exhibited by different instances of the macromolecule observed in a cryo-EM experiment, each of which is recorded as a particle image. The range of conformations and the conformation of each particle are not known a priori; one of the great promises of cryo-EM is to map this conformation space. Remarkable progress has been made in determining rigid structures from homogeneous samples of molecules in spite of the unknown orientation of each particle image and significant progress has been made in recovering a few distinct states from mixtures of rather distinct conformations, but more complex heterogeneous samples remain a major challenge. We introduce the ``hyper-molecule'' framework for modeling structures across different states of heterogeneous molecules, including continuums of states. The key idea behind this framework is representing heterogeneous macromolecules as high-dimensional objects, with the additional dimensions representing the conformation space. This idea is then refined to model properties such as localized heterogeneity. In addition, we introduce an algorithmic framework for recovering such maps of heterogeneous objects from experimental data using a Bayesian formulation of the problem and Markov chain Monte Carlo (MCMC) algorithms to address the computational challenges in recovering these high dimensional hyper-molecules. We demonstrate these ideas in a prototype applied to synthetic data.
Cryo-EM reconstruction of continuous heterogeneity by Laplacian spectral volumes
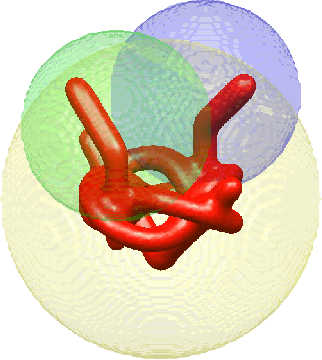
Jul 01, 2019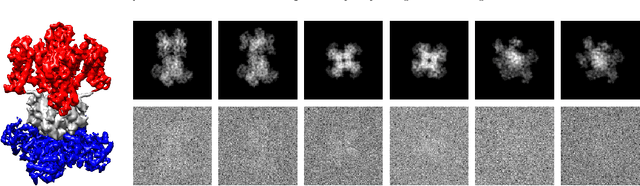
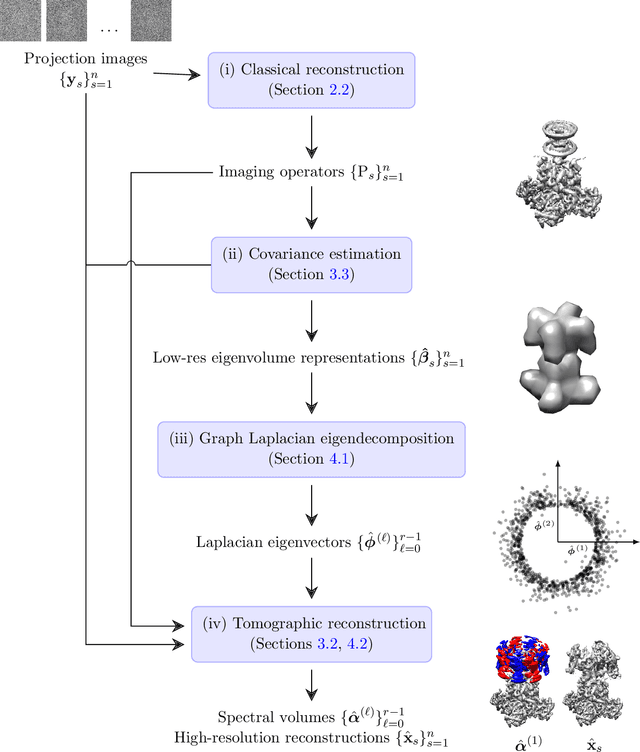
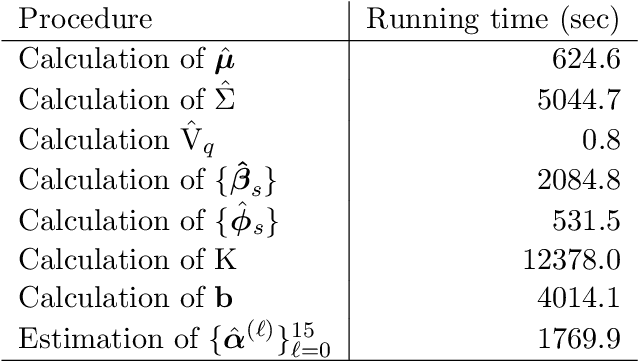
Abstract:Single-particle electron cryomicroscopy is an essential tool for high-resolution 3D reconstruction of proteins and other biological macromolecules. An important challenge in cryo-EM is the reconstruction of non-rigid molecules with parts that move and deform. Traditional reconstruction methods fail in these cases, resulting in smeared reconstructions of the moving parts. This poses a major obstacle for structural biologists, who need high-resolution reconstructions of entire macromolecules, moving parts included. To address this challenge, we present a new method for the reconstruction of macromolecules exhibiting continuous heterogeneity. The proposed method uses projection images from multiple viewing directions to construct a graph Laplacian through which the manifold of three-dimensional conformations is analyzed. The 3D molecular structures are then expanded in a basis of Laplacian eigenvectors, using a novel generalized tomographic reconstruction algorithm to compute the expansion coefficients. These coefficients, which we name spectral volumes, provide a high-resolution visualization of the molecular dynamics. We provide a theoretical analysis and evaluate the method empirically on several simulated data sets.
Kymatio: Scattering Transforms in Python
Dec 28, 2018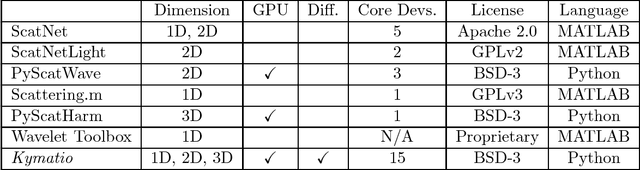
Abstract:The wavelet scattering transform is an invariant signal representation suitable for many signal processing and machine learning applications. We present the Kymatio software package, an easy-to-use, high-performance Python implementation of the scattering transform in 1D, 2D, and 3D that is compatible with modern deep learning frameworks. All transforms may be executed on a GPU (in addition to CPU), offering a considerable speed up over CPU implementations. The package also has a small memory footprint, resulting inefficient memory usage. The source code, documentation, and examples are available undera BSD license at https://www.kymat.io/
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge