Mahmoud Salem
Enabling High-Sparsity Foundational Llama Models with Efficient Pretraining and Deployment
May 06, 2024Abstract:Large language models (LLMs) have revolutionized Natural Language Processing (NLP), but their size creates computational bottlenecks. We introduce a novel approach to create accurate, sparse foundational versions of performant LLMs that achieve full accuracy recovery for fine-tuning tasks at up to 70% sparsity. We achieve this for the LLaMA-2 7B model by combining the SparseGPT one-shot pruning method and sparse pretraining of those models on a subset of the SlimPajama dataset mixed with a Python subset of The Stack dataset. We exhibit training acceleration due to sparsity on Cerebras CS-3 chips that closely matches theoretical scaling. In addition, we establish inference acceleration of up to 3x on CPUs by utilizing Neural Magic's DeepSparse engine and 1.7x on GPUs through Neural Magic's nm-vllm engine. The above gains are realized via sparsity alone, thus enabling further gains through additional use of quantization. Specifically, we show a total speedup on CPUs for sparse-quantized LLaMA models of up to 8.6x. We demonstrate these results across diverse, challenging tasks, including chat, instruction following, code generation, arithmetic reasoning, and summarization to prove their generality. This work paves the way for rapidly creating smaller and faster LLMs without sacrificing accuracy.
MediSwift: Efficient Sparse Pre-trained Biomedical Language Models
Mar 01, 2024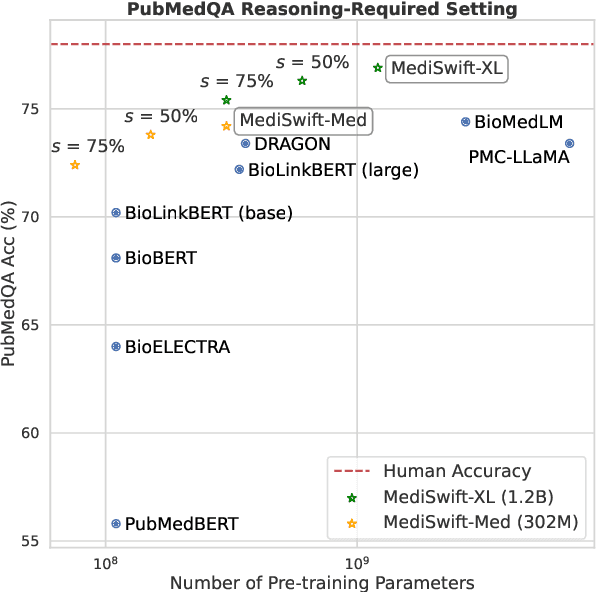
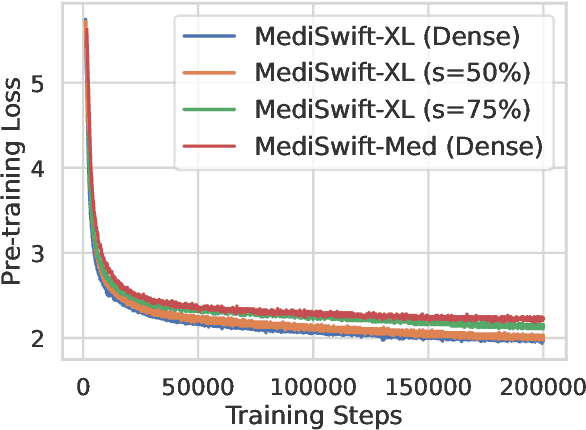
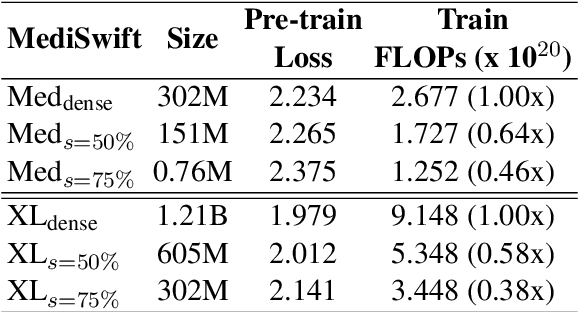
Abstract:Large language models (LLMs) are typically trained on general source data for various domains, but a recent surge in domain-specific LLMs has shown their potential to outperform general-purpose models in domain-specific tasks (e.g., biomedicine). Although domain-specific pre-training enhances efficiency and leads to smaller models, the computational costs of training these LLMs remain high, posing budgeting challenges. We introduce MediSwift, a suite of biomedical LMs that leverage sparse pre-training on domain-specific biomedical text data. By inducing up to 75% weight sparsity during the pre-training phase, MediSwift achieves a 2-2.5x reduction in training FLOPs. Notably, all sparse pre-training was performed on the Cerebras CS-2 system, which is specifically designed to realize the acceleration benefits from unstructured weight sparsity, thereby significantly enhancing the efficiency of the MediSwift models. Through subsequent dense fine-tuning and strategic soft prompting, MediSwift models outperform existing LLMs up to 7B parameters on biomedical tasks, setting new benchmarks w.r.t efficiency-accuracy on tasks such as PubMedQA. Our results show that sparse pre-training, along with dense fine-tuning and soft prompting, offers an effective method for creating high-performing, computationally efficient models in specialized domains.
Affective Visual Dialog: A Large-Scale Benchmark for Emotional Reasoning Based on Visually Grounded Conversations
Sep 12, 2023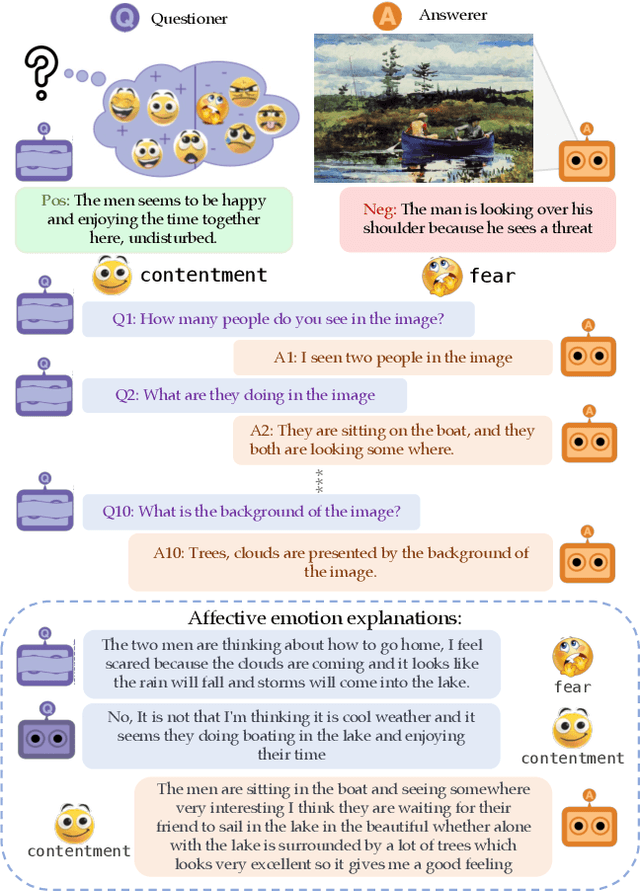
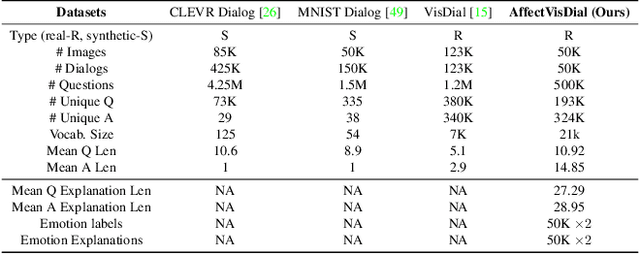

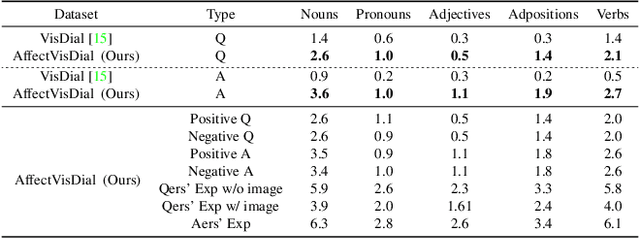
Abstract:We introduce Affective Visual Dialog, an emotion explanation and reasoning task as a testbed for research on understanding the formation of emotions in visually grounded conversations. The task involves three skills: (1) Dialog-based Question Answering (2) Dialog-based Emotion Prediction and (3) Affective emotion explanation generation based on the dialog. Our key contribution is the collection of a large-scale dataset, dubbed AffectVisDial, consisting of 50K 10-turn visually grounded dialogs as well as concluding emotion attributions and dialog-informed textual emotion explanations, resulting in a total of 27,180 working hours. We explain our design decisions in collecting the dataset and introduce the questioner and answerer tasks that are associated with the participants in the conversation. We train and demonstrate solid Affective Visual Dialog baselines adapted from state-of-the-art models. Remarkably, the responses generated by our models show promising emotional reasoning abilities in response to visually grounded conversations. Our project page is available at https://affective-visual-dialog.github.io.
Gumbel-Softmax Selective Networks
Nov 19, 2022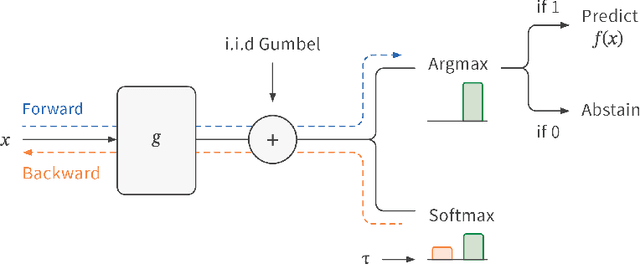
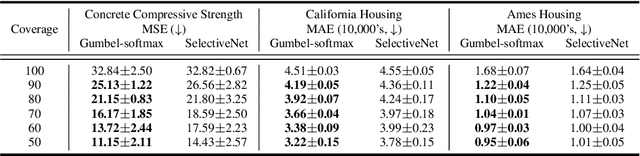

Abstract:ML models often operate within the context of a larger system that can adapt its response when the ML model is uncertain, such as falling back on safe defaults or a human in the loop. This commonly encountered operational context calls for principled techniques for training ML models with the option to abstain from predicting when uncertain. Selective neural networks are trained with an integrated option to abstain, allowing them to learn to recognize and optimize for the subset of the data distribution for which confident predictions can be made. However, optimizing selective networks is challenging due to the non-differentiability of the binary selection function (the discrete decision of whether to predict or abstain). This paper presents a general method for training selective networks that leverages the Gumbel-softmax reparameterization trick to enable selection within an end-to-end differentiable training framework. Experiments on public datasets demonstrate the potential of Gumbel-softmax selective networks for selective regression and classification.
Bounding generalization error with input compression: An empirical study with infinite-width networks
Jul 19, 2022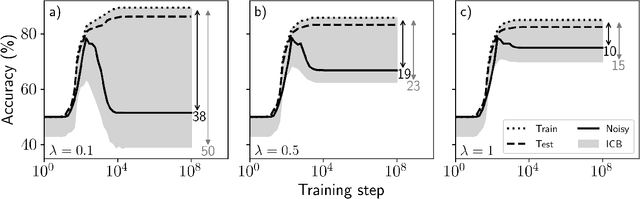

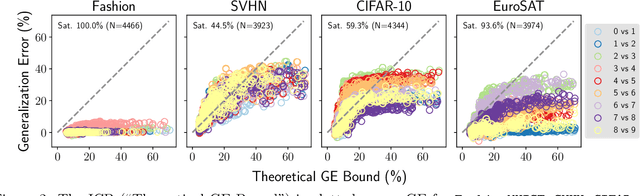
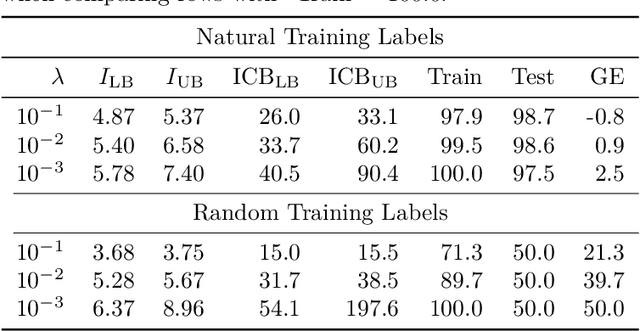
Abstract:Estimating the Generalization Error (GE) of Deep Neural Networks (DNNs) is an important task that often relies on availability of held-out data. The ability to better predict GE based on a single training set may yield overarching DNN design principles to reduce a reliance on trial-and-error, along with other performance assessment advantages. In search of a quantity relevant to GE, we investigate the Mutual Information (MI) between the input and final layer representations, using the infinite-width DNN limit to bound MI. An existing input compression-based GE bound is used to link MI and GE. To the best of our knowledge, this represents the first empirical study of this bound. In our attempt to empirically falsify the theoretical bound, we find that it is often tight for best-performing models. Furthermore, it detects randomization of training labels in many cases, reflects test-time perturbation robustness, and works well given only few training samples. These results are promising given that input compression is broadly applicable where MI can be estimated with confidence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge