Kevin Leong
Enabling High-Sparsity Foundational Llama Models with Efficient Pretraining and Deployment
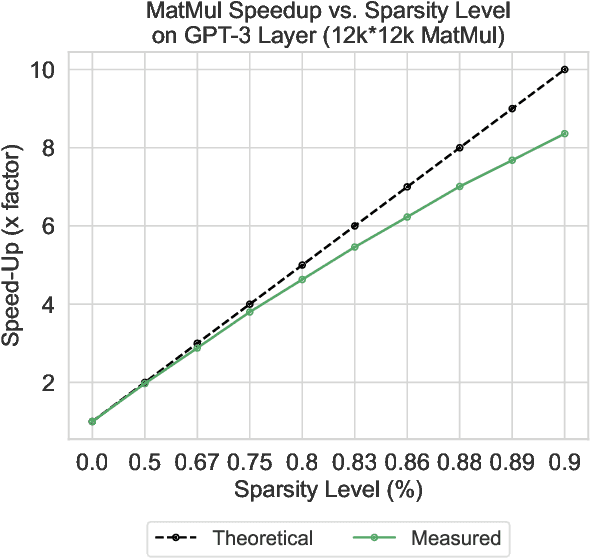
May 06, 2024Abstract:Large language models (LLMs) have revolutionized Natural Language Processing (NLP), but their size creates computational bottlenecks. We introduce a novel approach to create accurate, sparse foundational versions of performant LLMs that achieve full accuracy recovery for fine-tuning tasks at up to 70% sparsity. We achieve this for the LLaMA-2 7B model by combining the SparseGPT one-shot pruning method and sparse pretraining of those models on a subset of the SlimPajama dataset mixed with a Python subset of The Stack dataset. We exhibit training acceleration due to sparsity on Cerebras CS-3 chips that closely matches theoretical scaling. In addition, we establish inference acceleration of up to 3x on CPUs by utilizing Neural Magic's DeepSparse engine and 1.7x on GPUs through Neural Magic's nm-vllm engine. The above gains are realized via sparsity alone, thus enabling further gains through additional use of quantization. Specifically, we show a total speedup on CPUs for sparse-quantized LLaMA models of up to 8.6x. We demonstrate these results across diverse, challenging tasks, including chat, instruction following, code generation, arithmetic reasoning, and summarization to prove their generality. This work paves the way for rapidly creating smaller and faster LLMs without sacrificing accuracy.
MediSwift: Efficient Sparse Pre-trained Biomedical Language Models
Mar 01, 2024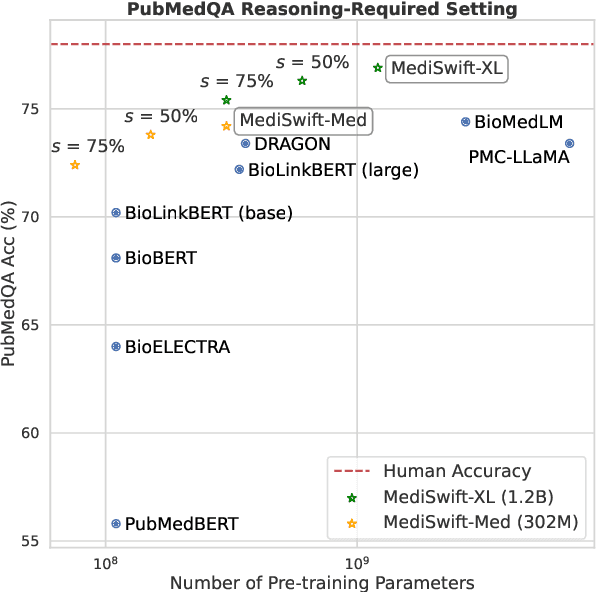
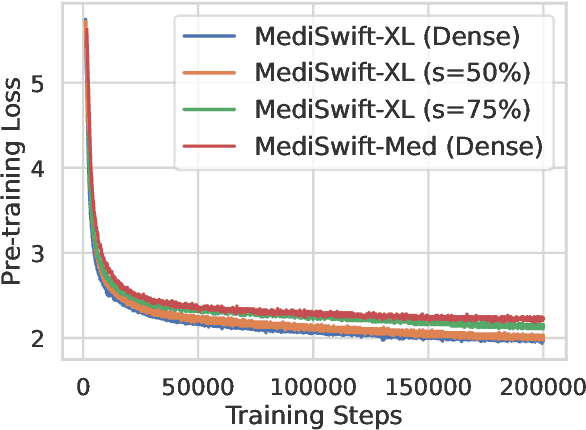
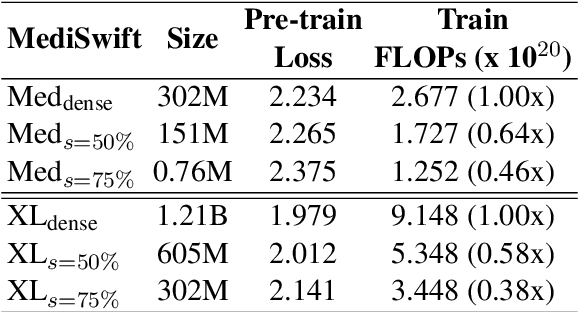
Abstract:Large language models (LLMs) are typically trained on general source data for various domains, but a recent surge in domain-specific LLMs has shown their potential to outperform general-purpose models in domain-specific tasks (e.g., biomedicine). Although domain-specific pre-training enhances efficiency and leads to smaller models, the computational costs of training these LLMs remain high, posing budgeting challenges. We introduce MediSwift, a suite of biomedical LMs that leverage sparse pre-training on domain-specific biomedical text data. By inducing up to 75% weight sparsity during the pre-training phase, MediSwift achieves a 2-2.5x reduction in training FLOPs. Notably, all sparse pre-training was performed on the Cerebras CS-2 system, which is specifically designed to realize the acceleration benefits from unstructured weight sparsity, thereby significantly enhancing the efficiency of the MediSwift models. Through subsequent dense fine-tuning and strategic soft prompting, MediSwift models outperform existing LLMs up to 7B parameters on biomedical tasks, setting new benchmarks w.r.t efficiency-accuracy on tasks such as PubMedQA. Our results show that sparse pre-training, along with dense fine-tuning and soft prompting, offers an effective method for creating high-performing, computationally efficient models in specialized domains.
SPDF: Sparse Pre-training and Dense Fine-tuning for Large Language Models
Mar 18, 2023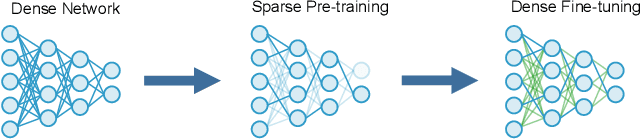

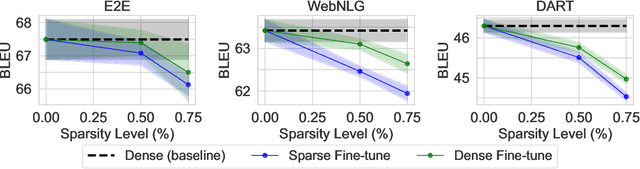
Abstract:The pre-training and fine-tuning paradigm has contributed to a number of breakthroughs in Natural Language Processing (NLP). Instead of directly training on a downstream task, language models are first pre-trained on large datasets with cross-domain knowledge (e.g., Pile, MassiveText, etc.) and then fine-tuned on task-specific data (e.g., natural language generation, text summarization, etc.). Scaling the model and dataset size has helped improve the performance of LLMs, but unfortunately, this also leads to highly prohibitive computational costs. Pre-training LLMs often require orders of magnitude more FLOPs than fine-tuning and the model capacity often remains the same between the two phases. To achieve training efficiency w.r.t training FLOPs, we propose to decouple the model capacity between the two phases and introduce Sparse Pre-training and Dense Fine-tuning (SPDF). In this work, we show the benefits of using unstructured weight sparsity to train only a subset of weights during pre-training (Sparse Pre-training) and then recover the representational capacity by allowing the zeroed weights to learn (Dense Fine-tuning). We demonstrate that we can induce up to 75% sparsity into a 1.3B parameter GPT-3 XL model resulting in a 2.5x reduction in pre-training FLOPs, without a significant loss in accuracy on the downstream tasks relative to the dense baseline. By rigorously evaluating multiple downstream tasks, we also establish a relationship between sparsity, task complexity, and dataset size. Our work presents a promising direction to train large GPT models at a fraction of the training FLOPs using weight sparsity while retaining the benefits of pre-trained textual representations for downstream tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge