Jingchi Jiang
GRAPHMOE: Amplifying Cognitive Depth of Mixture-of-Experts Network via Introducing Self-Rethinking Mechanism
Jan 14, 2025



Abstract:Traditional Mixture-of-Experts (MoE) networks benefit from utilizing multiple smaller expert models as opposed to a single large network. However, these experts typically operate independently, leaving a question open about whether interconnecting these models could enhance the performance of MoE networks. In response, we introduce GRAPHMOE, a novel method aimed at augmenting the cognitive depth of language models via a self-rethinking mechanism constructed on Pseudo GraphMoE networks. GRAPHMOE employs a recurrent routing strategy to simulate iterative thinking steps, thereby facilitating the flow of information among expert nodes. We implement the GRAPHMOE architecture using Low-Rank Adaptation techniques (LoRA) and conduct extensive experiments on various benchmark datasets. The experimental results reveal that GRAPHMOE outperforms other LoRA based models, achieving state-of-the-art (SOTA) performance. Additionally, this study explores a novel recurrent routing strategy that may inspire further advancements in enhancing the reasoning capabilities of language models.
Causal Discovery from Time-Series Data with Short-Term Invariance-Based Convolutional Neural Networks
Aug 15, 2024Abstract:Causal discovery from time-series data aims to capture both intra-slice (contemporaneous) and inter-slice (time-lagged) causality between variables within the temporal chain, which is crucial for various scientific disciplines. Compared to causal discovery from non-time-series data, causal discovery from time-series data necessitates more serialized samples with a larger amount of observed time steps. To address the challenges, we propose a novel gradient-based causal discovery approach STIC, which focuses on \textbf{S}hort-\textbf{T}erm \textbf{I}nvariance using \textbf{C}onvolutional neural networks to uncover the causal relationships from time-series data. Specifically, STIC leverages both the short-term time and mechanism invariance of causality within each window observation, which possesses the property of independence, to enhance sample efficiency. Furthermore, we construct two causal convolution kernels, which correspond to the short-term time and mechanism invariance respectively, to estimate the window causal graph. To demonstrate the necessity of convolutional neural networks for causal discovery from time-series data, we theoretically derive the equivalence between convolution and the underlying generative principle of time-series data under the assumption that the additive noise model is identifiable. Experimental evaluations conducted on both synthetic and FMRI benchmark datasets demonstrate that our STIC outperforms baselines significantly and achieves the state-of-the-art performance, particularly when the datasets contain a limited number of observed time steps. Code is available at \url{https://github.com/HITshenrj/STIC}.
ProSpec RL: Plan Ahead, then Execute
Jul 31, 2024Abstract:Imagining potential outcomes of actions before execution helps agents make more informed decisions, a prospective thinking ability fundamental to human cognition. However, mainstream model-free Reinforcement Learning (RL) methods lack the ability to proactively envision future scenarios, plan, and guide strategies. These methods typically rely on trial and error to adjust policy functions, aiming to maximize cumulative rewards or long-term value, even if such high-reward decisions place the environment in extremely dangerous states. To address this, we propose the Prospective (ProSpec) RL method, which makes higher-value, lower-risk optimal decisions by imagining future n-stream trajectories. Specifically, ProSpec employs a dynamic model to predict future states (termed "imagined states") based on the current state and a series of sampled actions. Furthermore, we integrate the concept of Model Predictive Control and introduce a cycle consistency constraint that allows the agent to evaluate and select the optimal actions from these trajectories. Moreover, ProSpec employs cycle consistency to mitigate two fundamental issues in RL: augmenting state reversibility to avoid irreversible events (low risk) and augmenting actions to generate numerous virtual trajectories, thereby improving data efficiency. We validated the effectiveness of our method on the DMControl benchmarks, where our approach achieved significant performance improvements. Code will be open-sourced upon acceptance.
FreqTSF: Time Series Forecasting Via Simulating Frequency Kramer-Kronig Relations
Jul 31, 2024Abstract:Time series forecasting (TSF) is immensely important in extensive applications, such as electricity transformation, financial trade, medical monitoring, and smart agriculture. Although Transformer-based methods can handle time series data, their ability to predict long-term time series is limited due to the ``anti-order" nature of the self-attention mechanism. To address this problem, we focus on frequency domain to weaken the impact of order in TSF and propose the FreqBlock, where we first obtain frequency representations through the Frequency Transform Module. Subsequently, a newly designed Frequency Cross Attention is used to obtian enhanced frequency representations between the real and imaginary parts, thus establishing a link between the attention mechanism and the inherent Kramer-Kronig relations (KKRs). Our backbone network, FreqTSF, adopts a residual structure by concatenating multiple FreqBlocks to simulate KKRs in the frequency domain and avoid degradation problems. On a theoretical level, we demonstrate that the proposed two modules can significantly reduce the time and memory complexity from $\mathcal{O}(L^2)$ to $\mathcal{O}(L)$ for each FreqBlock computation. Empirical studies on four benchmark datasets show that FreqTSF achieves an overall relative MSE reduction of 15\% and an overall relative MAE reduction of 11\% compared to the state-of-the-art methods. The code will be available soon.
Causal prompting model-based offline reinforcement learning
Jun 03, 2024Abstract:Model-based offline Reinforcement Learning (RL) allows agents to fully utilise pre-collected datasets without requiring additional or unethical explorations. However, applying model-based offline RL to online systems presents challenges, primarily due to the highly suboptimal (noise-filled) and diverse nature of datasets generated by online systems. To tackle these issues, we introduce the Causal Prompting Reinforcement Learning (CPRL) framework, designed for highly suboptimal and resource-constrained online scenarios. The initial phase of CPRL involves the introduction of the Hidden-Parameter Block Causal Prompting Dynamic (Hip-BCPD) to model environmental dynamics. This approach utilises invariant causal prompts and aligns hidden parameters to generalise to new and diverse online users. In the subsequent phase, a single policy is trained to address multiple tasks through the amalgamation of reusable skills, circumventing the need for training from scratch. Experiments conducted across datasets with varying levels of noise, including simulation-based and real-world offline datasets from the Dnurse APP, demonstrate that our proposed method can make robust decisions in out-of-distribution and noisy environments, outperforming contemporary algorithms. Additionally, we separately verify the contributions of Hip-BCPDs and the skill-reuse strategy to the robustness of performance. We further analyse the visualised structure of Hip-BCPD and the interpretability of sub-skills. We released our source code and the first ever real-world medical dataset for precise medical decision-making tasks.
Blood Glucose Control Via Pre-trained Counterfactual Invertible Neural Networks
May 23, 2024



Abstract:Type 1 diabetes mellitus (T1D) is characterized by insulin deficiency and blood glucose (BG) control issues. The state-of-the-art solution for continuous BG control is reinforcement learning (RL), where an agent can dynamically adjust exogenous insulin doses in time to maintain BG levels within the target range. However, due to the lack of action guidance, the agent often needs to learn from randomized trials to understand misleading correlations between exogenous insulin doses and BG levels, which can lead to instability and unsafety. To address these challenges, we propose an introspective RL based on Counterfactual Invertible Neural Networks (CINN). We use the pre-trained CINN as a frozen introspective block of the RL agent, which integrates forward prediction and counterfactual inference to guide the policy updates, promoting more stable and safer BG control. Constructed based on interpretable causal order, CINN employs bidirectional encoders with affine coupling layers to ensure invertibility while using orthogonal weight normalization to enhance the trainability, thereby ensuring the bidirectional differentiability of network parameters. We experimentally validate the accuracy and generalization ability of the pre-trained CINN in BG prediction and counterfactual inference for action. Furthermore, our experimental results highlight the effectiveness of pre-trained CINN in guiding RL policy updates for more accurate and safer BG control.
Causal Coupled Mechanisms: A Control Method with Cooperation and Competition for Complex System
Sep 15, 2022



Abstract:Complex systems are ubiquitous in the real world and tend to have complicated and poorly understood dynamics. For their control issues, the challenge is to guarantee accuracy, robustness, and generalization in such bloated and troubled environments. Fortunately, a complex system can be divided into multiple modular structures that human cognition appears to exploit. Inspired by this cognition, a novel control method, Causal Coupled Mechanisms (CCMs), is proposed that explores the cooperation in division and competition in combination. Our method employs the theory of hierarchical reinforcement learning (HRL), in which 1) the high-level policy with competitive awareness divides the whole complex system into multiple functional mechanisms, and 2) the low-level policy finishes the control task of each mechanism. Specifically for cooperation, a cascade control module helps the series operation of CCMs, and a forward coupled reasoning module is used to recover the coupling information lost in the division process. On both synthetic systems and a real-world biological regulatory system, the CCM method achieves robust and state-of-the-art control results even with unpredictable random noise. Moreover, generalization results show that reusing prepared specialized CCMs helps to perform well in environments with different confounders and dynamics.
Medical Knowledge Embedding Based on Recursive Neural Network for Multi-Disease Diagnosis
Sep 22, 2018



Abstract:The representation of knowledge based on first-order logic captures the richness of natural language and supports multiple probabilistic inference models. Although symbolic representation enables quantitative reasoning with statistical probability, it is difficult to utilize with machine learning models as they perform numerical operations. In contrast, knowledge embedding (i.e., high-dimensional and continuous vectors) is a feasible approach to complex reasoning that can not only retain the semantic information of knowledge but also establish the quantifiable relationship among them. In this paper, we propose recursive neural knowledge network (RNKN), which combines medical knowledge based on first-order logic with recursive neural network for multi-disease diagnosis. After RNKN is efficiently trained from manually annotated Chinese Electronic Medical Records (CEMRs), diagnosis-oriented knowledge embeddings and weight matrixes are learned. Experimental results verify that the diagnostic accuracy of RNKN is superior to that of some classical machine learning models and Markov logic network (MLN). The results also demonstrate that the more explicit the evidence extracted from CEMRs is, the better is the performance achieved. RNKN gradually exhibits the interpretation of knowledge embeddings as the number of training epochs increases.
De-identification of medical records using conditional random fields and long short-term memory networks
Sep 29, 2017
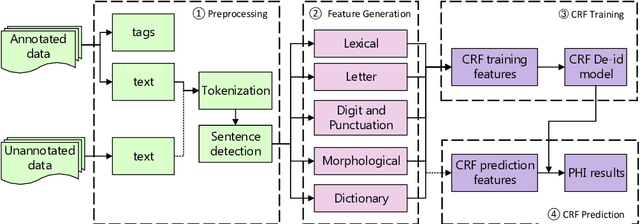

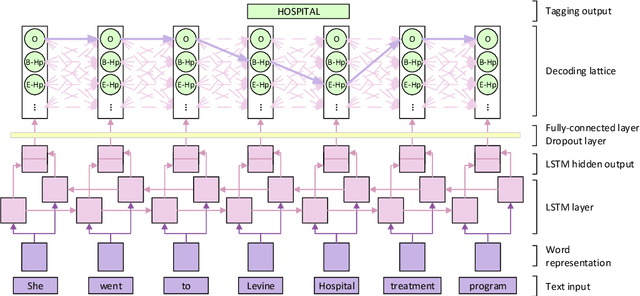
Abstract:The CEGS N-GRID 2016 Shared Task 1 in Clinical Natural Language Processing focuses on the de-identification of psychiatric evaluation records. This paper describes two participating systems of our team, based on conditional random fields (CRFs) and long short-term memory networks (LSTMs). A pre-processing module was introduced for sentence detection and tokenization before de-identification. For CRFs, manually extracted rich features were utilized to train the model. For LSTMs, a character-level bi-directional LSTM network was applied to represent tokens and classify tags for each token, following which a decoding layer was stacked to decode the most probable protected health information (PHI) terms. The LSTM-based system attained an i2b2 strict micro-F_1 measure of 89.86%, which was higher than that of the CRF-based system.
EMR-based medical knowledge representation and inference via Markov random fields and distributed representation learning
Sep 20, 2017



Abstract:Objective: Electronic medical records (EMRs) contain an amount of medical knowledge which can be used for clinical decision support (CDS). Our objective is a general system that can extract and represent these knowledge contained in EMRs to support three CDS tasks: test recommendation, initial diagnosis, and treatment plan recommendation, with the given condition of one patient. Methods: We extracted four kinds of medical entities from records and constructed an EMR-based medical knowledge network (EMKN), in which nodes are entities and edges reflect their co-occurrence in a single record. Three bipartite subgraphs (bi-graphs) were extracted from the EMKN to support each task. One part of the bi-graph was the given condition (e.g., symptoms), and the other was the condition to be inferred (e.g., diseases). Each bi-graph was regarded as a Markov random field to support the inference. Three lazy energy functions and one parameter-based energy function were proposed, as well as two knowledge representation learning-based energy functions, which can provide a distributed representation of medical entities. Three measures were utilized for performance evaluation. Results: On the initial diagnosis task, 80.11% of the test records identified at least one correct disease from top 10 candidates. Test and treatment recommendation results were 87.88% and 92.55%, respectively. These results altogether indicate that the proposed system outperformed the baseline methods. The distributed representation of medical entities does reflect similarity relationships in regards to knowledge level. Conclusion: Combining EMKN and MRF is an effective approach for general medical knowledge representation and inference. Different tasks, however, require designing their energy functions individually.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge