Bo Lv
HyCoRA: Hyper-Contrastive Role-Adaptive Learning for Role-Playing
Nov 11, 2025Abstract:Multi-character role-playing aims to equip models with the capability to simulate diverse roles. Existing methods either use one shared parameterized module across all roles or assign a separate parameterized module to each role. However, the role-shared module may ignore distinct traits of each role, weakening personality learning, while the role-specific module may overlook shared traits across multiple roles, hindering commonality modeling. In this paper, we propose a novel HyCoRA: Hyper-Contrastive Role-Adaptive learning framework, which efficiently improves multi-character role-playing ability by balancing the learning of distinct and shared traits. Specifically, we propose a Hyper-Half Low-Rank Adaptation structure, where one half is a role-specific module generated by a lightweight hyper-network, and the other half is a trainable role-shared module. The role-specific module is devised to represent distinct persona signatures, while the role-shared module serves to capture common traits. Moreover, to better reflect distinct personalities across different roles, we design a hyper-contrastive learning mechanism to help the hyper-network distinguish their unique characteristics. Extensive experimental results on both English and Chinese available benchmarks demonstrate the superiority of our framework. Further GPT-4 evaluations and visual analyses also verify the capability of HyCoRA to capture role characteristics.
GRAPHMOE: Amplifying Cognitive Depth of Mixture-of-Experts Network via Introducing Self-Rethinking Mechanism
Jan 14, 2025



Abstract:Traditional Mixture-of-Experts (MoE) networks benefit from utilizing multiple smaller expert models as opposed to a single large network. However, these experts typically operate independently, leaving a question open about whether interconnecting these models could enhance the performance of MoE networks. In response, we introduce GRAPHMOE, a novel method aimed at augmenting the cognitive depth of language models via a self-rethinking mechanism constructed on Pseudo GraphMoE networks. GRAPHMOE employs a recurrent routing strategy to simulate iterative thinking steps, thereby facilitating the flow of information among expert nodes. We implement the GRAPHMOE architecture using Low-Rank Adaptation techniques (LoRA) and conduct extensive experiments on various benchmark datasets. The experimental results reveal that GRAPHMOE outperforms other LoRA based models, achieving state-of-the-art (SOTA) performance. Additionally, this study explores a novel recurrent routing strategy that may inspire further advancements in enhancing the reasoning capabilities of language models.
SpecFuse: Ensembling Large Language Models via Next-Segment Prediction
Dec 10, 2024



Abstract:Ensembles of generative large language models (LLMs) can integrate the strengths of different LLMs to compensate for the limitations of individual models. However, recent work has focused on training an additional fusion model to combine complete responses from multiple LLMs, failing to tap into their collaborative potential to generate higher-quality responses. Moreover, as the additional fusion model is trained on a specialized dataset, these methods struggle with generalizing to open-domain queries from online users. In this paper, we propose SpecFuse, a novel ensemble framework that outputs the fused result by iteratively producing the next segment through collaboration among LLMs. This is achieved through cyclic execution of its inference and verification components. In each round, the inference component invokes each base LLM to generate candidate segments in parallel, and the verify component calls these LLMs again to predict the ranking of the segments. The top-ranked segment is then broadcast to all LLMs, encouraging them to generate higher-quality segments in the next round. This approach also allows the base LLMs to be plug-and-play, without any training or adaptation, avoiding generalization limitations. Furthermore, to conserve computational resources, we propose a model exit mechanism that dynamically excludes models exhibiting poor performance in previous rounds during each query response. In this way, it effectively reduces the number of model calls while maintaining overall performance.
KVPruner: Structural Pruning for Faster and Memory-Efficient Large Language Models
Sep 17, 2024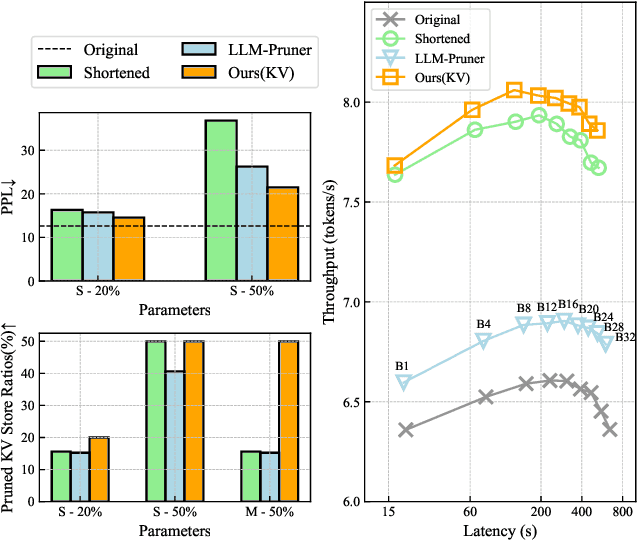
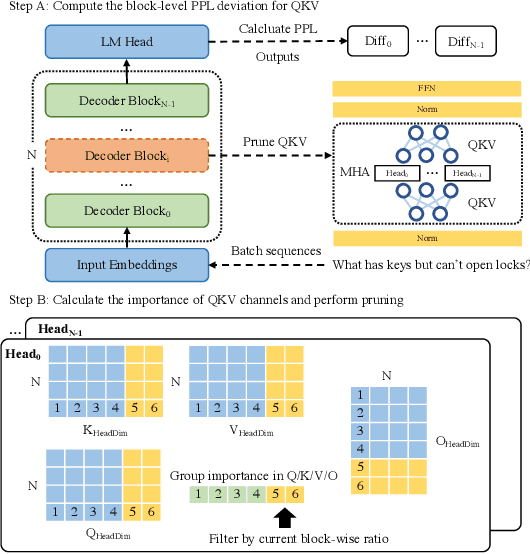
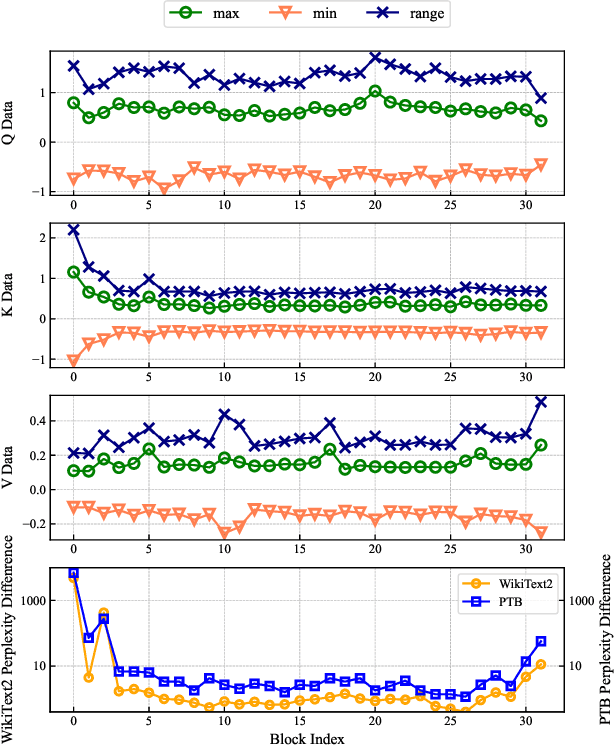
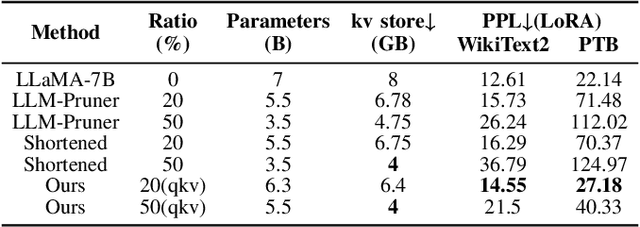
Abstract:The bottleneck associated with the key-value(KV) cache presents a significant challenge during the inference processes of large language models. While depth pruning accelerates inference, it requires extensive recovery training, which can take up to two weeks. On the other hand, width pruning retains much of the performance but offers slight speed gains. To tackle these challenges, we propose KVPruner to improve model efficiency while maintaining performance. Our method uses global perplexity-based analysis to determine the importance ratio for each block and provides multiple strategies to prune non-essential KV channels within blocks. Compared to the original model, KVPruner reduces runtime memory usage by 50% and boosts throughput by over 35%. Additionally, our method requires only two hours of LoRA fine-tuning on small datasets to recover most of the performance.
BioMNER: A Dataset for Biomedical Method Entity Recognition
Jun 28, 2024Abstract:Named entity recognition (NER) stands as a fundamental and pivotal task within the realm of Natural Language Processing. Particularly within the domain of Biomedical Method NER, this task presents notable challenges, stemming from the continual influx of domain-specific terminologies in scholarly literature. Current research in Biomedical Method (BioMethod) NER suffers from a scarcity of resources, primarily attributed to the intricate nature of methodological concepts, which necessitate a profound understanding for precise delineation. In this study, we propose a novel dataset for biomedical method entity recognition, employing an automated BioMethod entity recognition and information retrieval system to assist human annotation. Furthermore, we comprehensively explore a range of conventional and contemporary open-domain NER methodologies, including the utilization of cutting-edge large-scale language models (LLMs) customised to our dataset. Our empirical findings reveal that the large parameter counts of language models surprisingly inhibit the effective assimilation of entity extraction patterns pertaining to biomedical methods. Remarkably, the approach, leveraging the modestly sized ALBERT model (only 11MB), in conjunction with conditional random fields (CRF), achieves state-of-the-art (SOTA) performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge