Jennifer Keegan
Adaptive Hierarchical Dual Consistency for Semi-Supervised Left Atrium Segmentation on Cross-Domain Data
Sep 20, 2021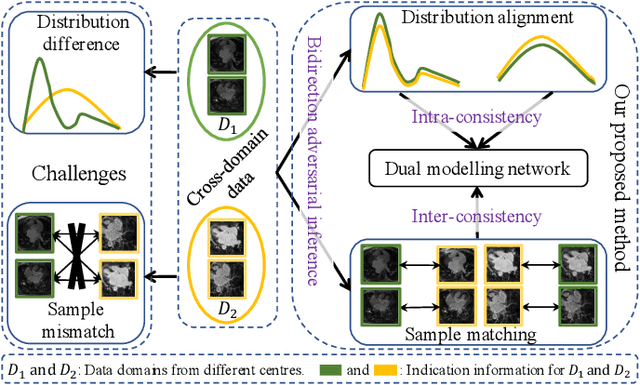
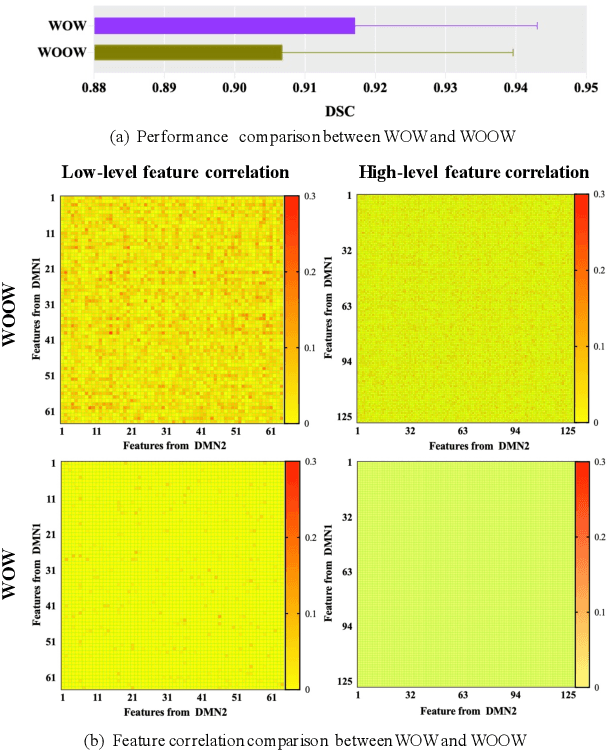
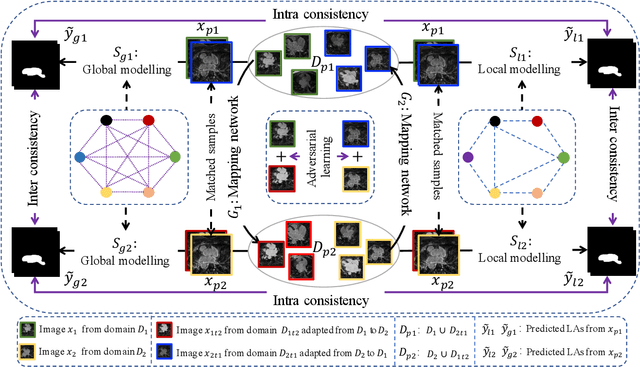
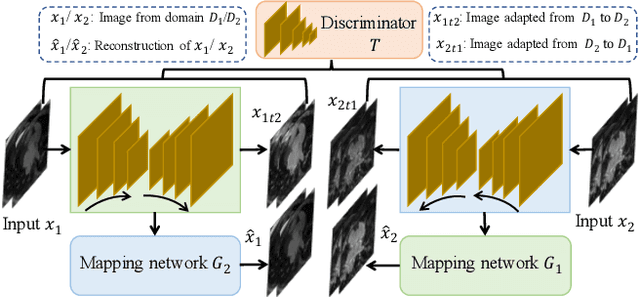
Abstract:Semi-supervised learning provides great significance in left atrium (LA) segmentation model learning with insufficient labelled data. Generalising semi-supervised learning to cross-domain data is of high importance to further improve model robustness. However, the widely existing distribution difference and sample mismatch between different data domains hinder the generalisation of semi-supervised learning. In this study, we alleviate these problems by proposing an Adaptive Hierarchical Dual Consistency (AHDC) for the semi-supervised LA segmentation on cross-domain data. The AHDC mainly consists of a Bidirectional Adversarial Inference module (BAI) and a Hierarchical Dual Consistency learning module (HDC). The BAI overcomes the difference of distributions and the sample mismatch between two different domains. It mainly learns two mapping networks adversarially to obtain two matched domains through mutual adaptation. The HDC investigates a hierarchical dual learning paradigm for cross-domain semi-supervised segmentation based on the obtained matched domains. It mainly builds two dual-modelling networks for mining the complementary information in both intra-domain and inter-domain. For the intra-domain learning, a consistency constraint is applied to the dual-modelling targets to exploit the complementary modelling information. For the inter-domain learning, a consistency constraint is applied to the LAs modelled by two dual-modelling networks to exploit the complementary knowledge among different data domains. We demonstrated the performance of our proposed AHDC on four 3D late gadolinium enhancement cardiac MR (LGE-CMR) datasets from different centres and a 3D CT dataset. Compared to other state-of-the-art methods, our proposed AHDC achieved higher segmentation accuracy, which indicated its capability in the cross-domain semi-supervised LA segmentation.
JAS-GAN: Generative Adversarial Network Based Joint Atrium and Scar Segmentations on Unbalanced Atrial Targets
May 01, 2021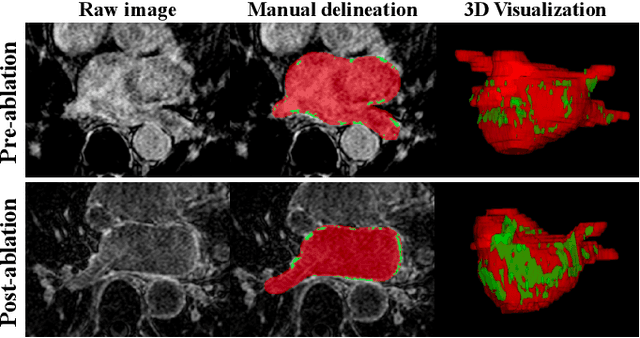
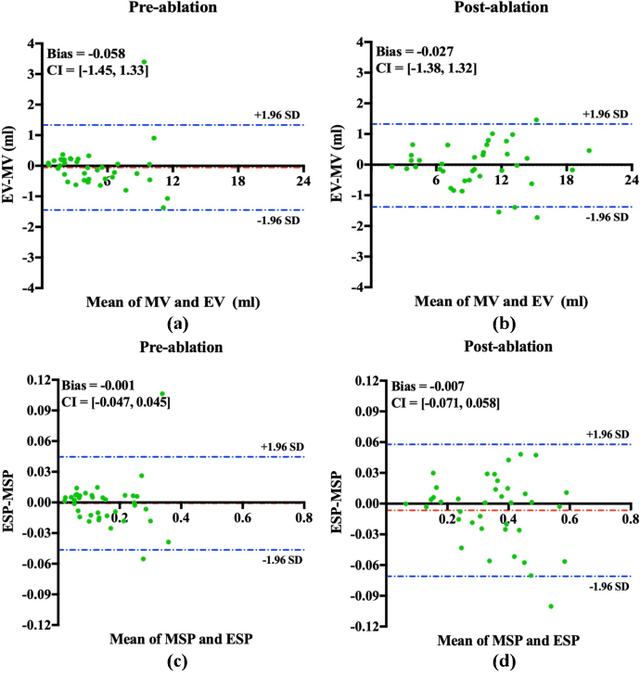
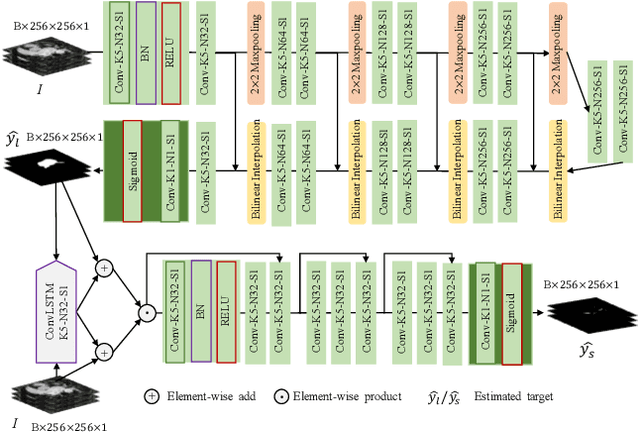
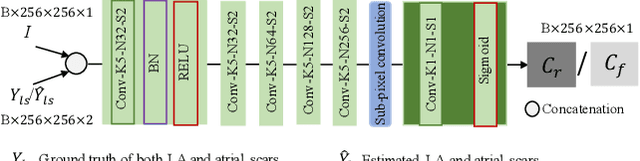
Abstract:Automated and accurate segmentations of left atrium (LA) and atrial scars from late gadolinium-enhanced cardiac magnetic resonance (LGE CMR) images are in high demand for quantifying atrial scars. The previous quantification of atrial scars relies on a two-phase segmentation for LA and atrial scars due to their large volume difference (unbalanced atrial targets). In this paper, we propose an inter-cascade generative adversarial network, namely JAS-GAN, to segment the unbalanced atrial targets from LGE CMR images automatically and accurately in an end-to-end way. Firstly, JAS-GAN investigates an adaptive attention cascade to automatically correlate the segmentation tasks of the unbalanced atrial targets. The adaptive attention cascade mainly models the inclusion relationship of the two unbalanced atrial targets, where the estimated LA acts as the attention map to adaptively focus on the small atrial scars roughly. Then, an adversarial regularization is applied to the segmentation tasks of the unbalanced atrial targets for making a consistent optimization. It mainly forces the estimated joint distribution of LA and atrial scars to match the real ones. We evaluated the performance of our JAS-GAN on a 3D LGE CMR dataset with 192 scans. Compared with the state-of-the-art methods, our proposed approach yielded better segmentation performance (Average Dice Similarity Coefficient (DSC) values of 0.946 and 0.821 for LA and atrial scars, respectively), which indicated the effectiveness of our proposed approach for segmenting unbalanced atrial targets.
Three-Dimensional Embedded Attentive RNN (3D-EAR) Segmentor for Left Ventricle Delineation from Myocardial Velocity Mapping
Apr 26, 2021
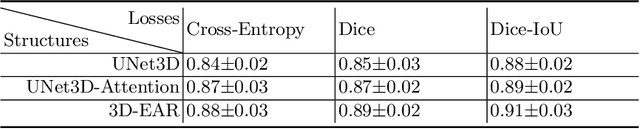
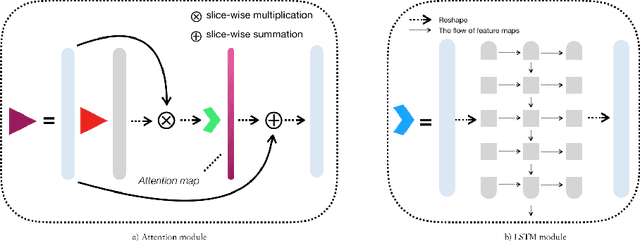
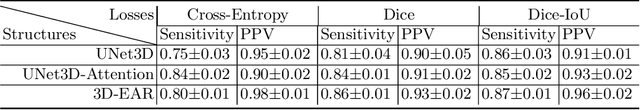
Abstract:Myocardial Velocity Mapping Cardiac MR (MVM-CMR) can be used to measure global and regional myocardial velocities with proved reproducibility. Accurate left ventricle delineation is a prerequisite for robust and reproducible myocardial velocity estimation. Conventional manual segmentation on this dataset can be time-consuming and subjective, and an effective fully automated delineation method is highly in demand. By leveraging recently proposed deep learning-based semantic segmentation approaches, in this study, we propose a novel fully automated framework incorporating a 3D-UNet backbone architecture with Embedded multichannel Attention mechanism and LSTM based Recurrent neural networks (RNN) for the MVM-CMR datasets (dubbed 3D-EAR segmentor). The proposed method also utilises the amalgamation of magnitude and phase images as input to realise an information fusion of this multichannel dataset and exploring the correlations of temporal frames via the embedded RNN. By comparing the baseline model of 3D-UNet and ablation studies with and without embedded attentive LSTM modules and various loss functions, we can demonstrate that the proposed model has outperformed the state-of-the-art baseline models with significant improvement.
Automated Multi-Channel Segmentation for the 4D Myocardial Velocity Mapping Cardiac MR
Dec 16, 2020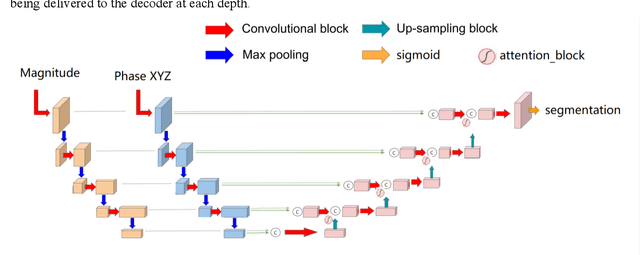
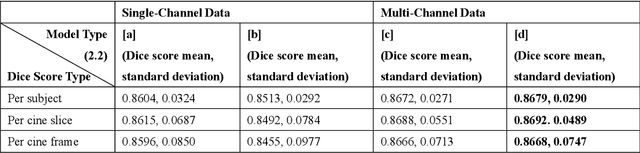

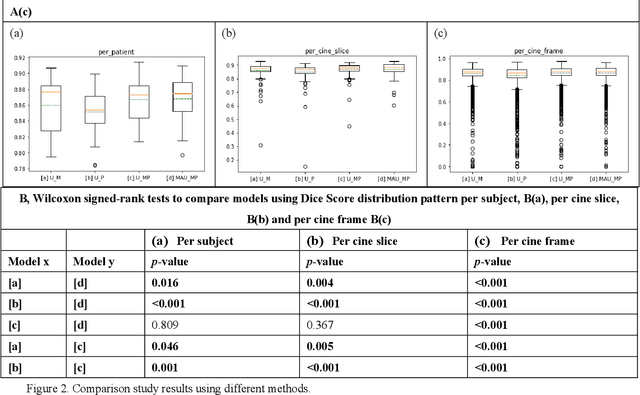
Abstract:Four-dimensional (4D) left ventricular myocardial velocity mapping (MVM) is a cardiac magnetic resonance (CMR) technique that allows assessment of cardiac motion in three orthogonal directions. Accurate and reproducible delineation of the myocardium is crucial for accurate analysis of peak systolic and diastolic myocardial velocities. In addition to the conventionally available magnitude CMR data, 4D MVM also acquires three velocity-encoded phase datasets which are used to generate velocity maps. These can be used to facilitate and improve myocardial delineation. Based on the success of deep learning in medical image processing, we propose a novel automated framework that improves the standard U-Net based methods on these CMR multi-channel data (magnitude and phase) by cross-channel fusion with attention module and shape information based post-processing to achieve accurate delineation of both epicardium and endocardium contours. To evaluate the results, we employ the widely used Dice scores and the quantification of myocardial longitudinal peak velocities. Our proposed network trained with multi-channel data shows enhanced performance compared to standard U-Net based networks trained with single-channel data. Based on the results, our method provides compelling evidence for the design and application for the multi-channel image analysis of the 4D MVM CMR data.
Simultaneous Left Atrium Anatomy and Scar Segmentations via Deep Learning in Multiview Information with Attention
Feb 02, 2020
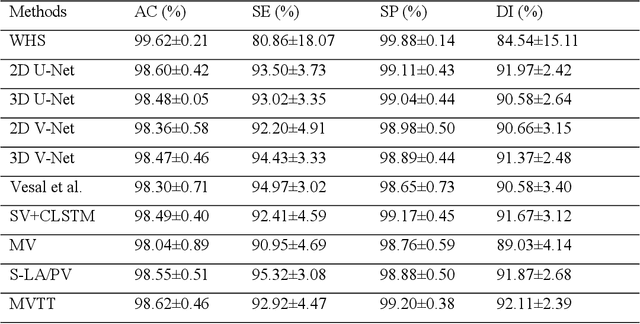

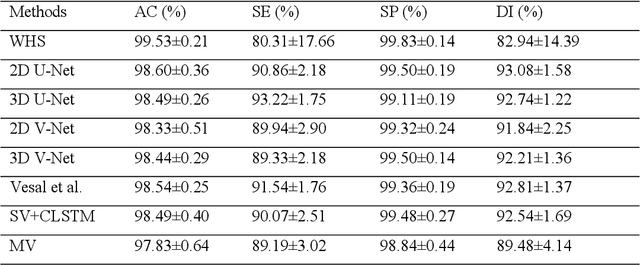
Abstract:Three-dimensional late gadolinium enhanced (LGE) cardiac MR (CMR) of left atrial scar in patients with atrial fibrillation (AF) has recently emerged as a promising technique to stratify patients, to guide ablation therapy and to predict treatment success. This requires a segmentation of the high intensity scar tissue and also a segmentation of the left atrium (LA) anatomy, the latter usually being derived from a separate bright-blood acquisition. Performing both segmentations automatically from a single 3D LGE CMR acquisition would eliminate the need for an additional acquisition and avoid subsequent registration issues. In this paper, we propose a joint segmentation method based on multiview two-task (MVTT) recursive attention model working directly on 3D LGE CMR images to segment the LA (and proximal pulmonary veins) and to delineate the scar on the same dataset. Using our MVTT recursive attention model, both the LA anatomy and scar can be segmented accurately (mean Dice score of 93% for the LA anatomy and 87% for the scar segmentations) and efficiently (~0.27 seconds to simultaneously segment the LA anatomy and scars directly from the 3D LGE CMR dataset with 60-68 2D slices). Compared to conventional unsupervised learning and other state-of-the-art deep learning based methods, the proposed MVTT model achieved excellent results, leading to an automatic generation of a patient-specific anatomical model combined with scar segmentation for patients in AF.
Discriminative Consistent Domain Generation for Semi-supervised Learning
Jul 24, 2019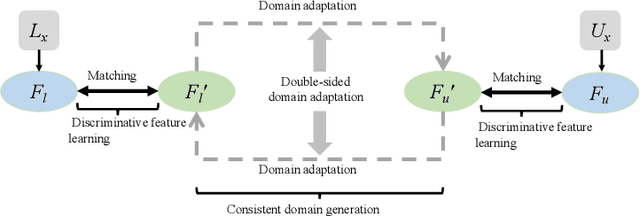
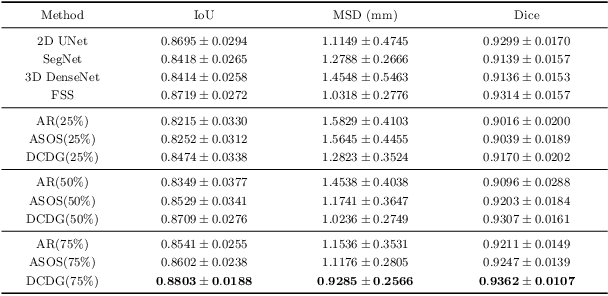
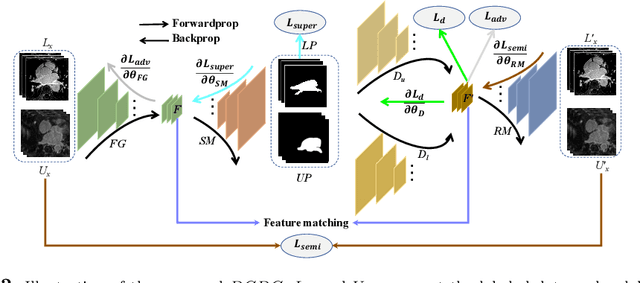
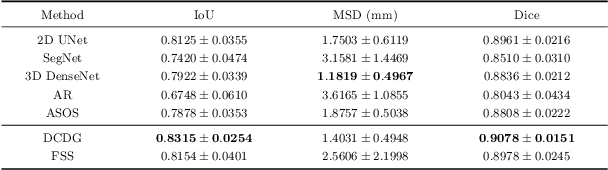
Abstract:Deep learning based task systems normally rely on a large amount of manually labeled training data, which is expensive to obtain and subject to operator variations. Moreover, it does not always hold that the manually labeled data and the unlabeled data are sitting in the same distribution. In this paper, we alleviate these problems by proposing a discriminative consistent domain generation (DCDG) approach to achieve a semi-supervised learning. The discriminative consistent domain is achieved by a double-sided domain adaptation. The double-sided domain adaptation aims to make a fusion of the feature spaces of labeled data and unlabeled data. In this way, we can fit the differences of various distributions between labeled data and unlabeled data. In order to keep the discriminativeness of generated consistent domain for the task learning, we apply an indirect learning for the double-sided domain adaptation. Based on the generated discriminative consistent domain, we can use the unlabeled data to learn the task model along with the labeled data via a consistent image generation. We demonstrate the performance of our proposed DCDG on the late gadolinium enhancement cardiac MRI (LGE-CMRI) images acquired from patients with atrial fibrillation in two clinical centers for the segmentation of the left atrium anatomy (LA) and proximal pulmonary veins (PVs). The experiments show that our semi-supervised approach achieves compelling segmentation results, which can prove the robustness of DCDG for the semi-supervised learning using the unlabeled data along with labeled data acquired from a single center or multicenter studies.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019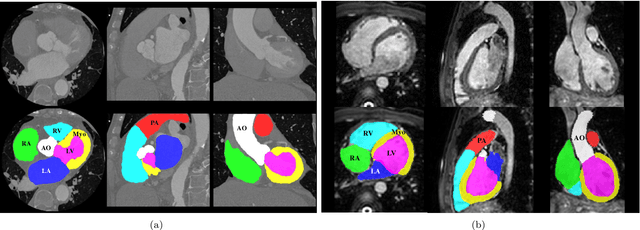
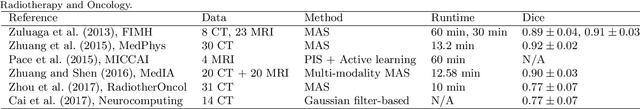
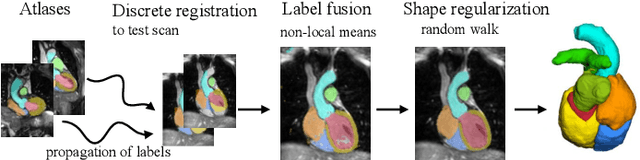
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
Atrial Scar Quantification via Multi-scale CNN in the Graph-cuts Framework
Feb 21, 2019
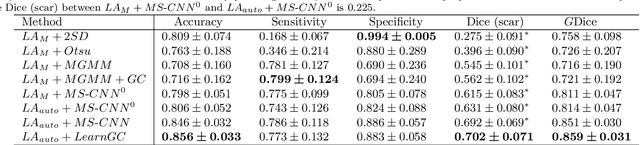
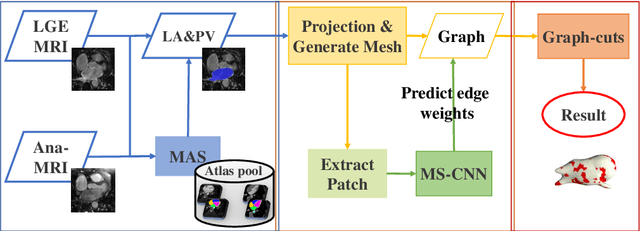

Abstract:Late gadolinium enhancement magnetic resonance imaging (LGE MRI) appears to be a promising alternative for scar assessment in patients with atrial fibrillation (AF). Automating the quantification and analysis of atrial scars can be challenging due to the low image quality. In this work, we propose a fully automated method based on the graph-cuts framework, where the potentials of the graph are learned on a surface mesh of the left atrium (LA) using a multi-scale convolutional neural network (MS-CNN). For validation, we have employed fifty-eight images with manual delineations. MS-CNN, which can efficiently incorporate both the local and global texture information of the images, has been shown to evidently improve the segmentation accuracy of the proposed graph-cuts based method. The segmentation could be further improved when the contribution between the t-link and n-link weights of the graph is balanced. The proposed method achieves a mean accuracy of 0.856 +- 0.033 and mean Dice score of 0.702 +- 0.071 for LA scar quantification. Compared with the conventional methods, which are based on the manual delineation of LA for initialization, our method is fully automatic and has demonstrated significantly better Dice score and accuracy (p < 0.01). The method is promising and can be useful in diagnosis and prognosis of AF.
Atrial fibrosis quantification based on maximum likelihood estimator of multivariate images
Oct 22, 2018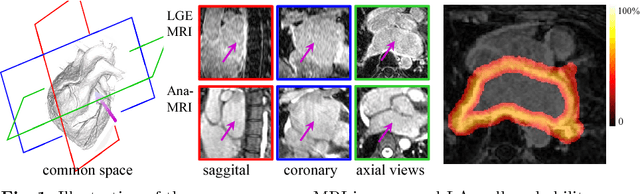
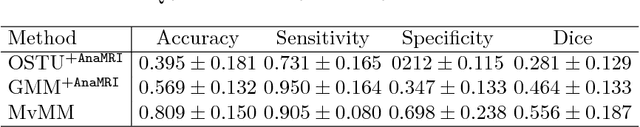
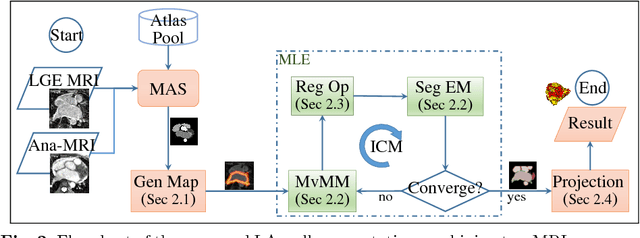
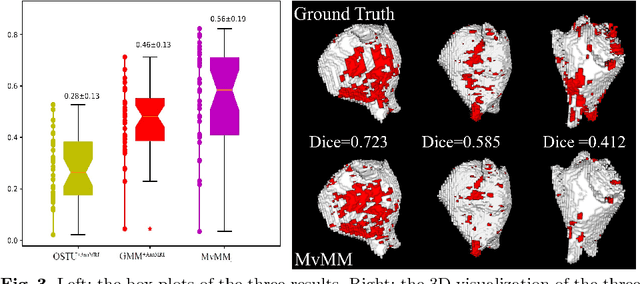
Abstract:We present a fully-automated segmentation and quantification of the left atrial (LA) fibrosis and scars combining two cardiac MRIs, one is the target late gadolinium-enhanced (LGE) image, and the other is an anatomical MRI from the same acquisition session. We formulate the joint distribution of images using a multivariate mixture model (MvMM), and employ the maximum likelihood estimator (MLE) for texture classification of the images simultaneously. The MvMM can also embed transformations assigned to the images to correct the misregistration. The iterated conditional mode algorithm is adopted for optimization. This method first extracts the anatomical shape of the LA, and then estimates a prior probability map. It projects the resulting segmentation onto the LA surface, for quantification and analysis of scarring. We applied the proposed method to 36 clinical data sets and obtained promising results (Accuracy: $0.809\pm .150$, Dice: $0.556\pm.187$). We compared the method with the conventional algorithms and showed an evidently and statistically better performance ($p<0.03$).
Translational Motion Compensation for Soft Tissue Velocity Images
Aug 20, 2018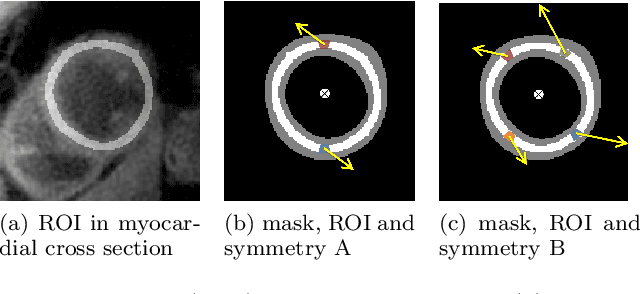
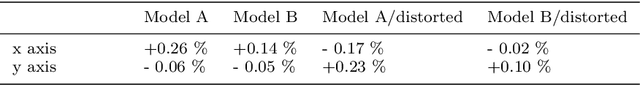
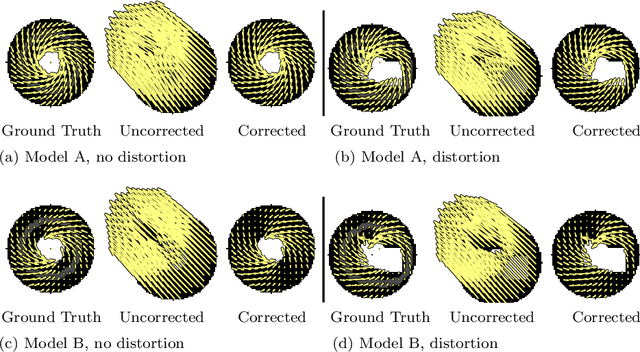
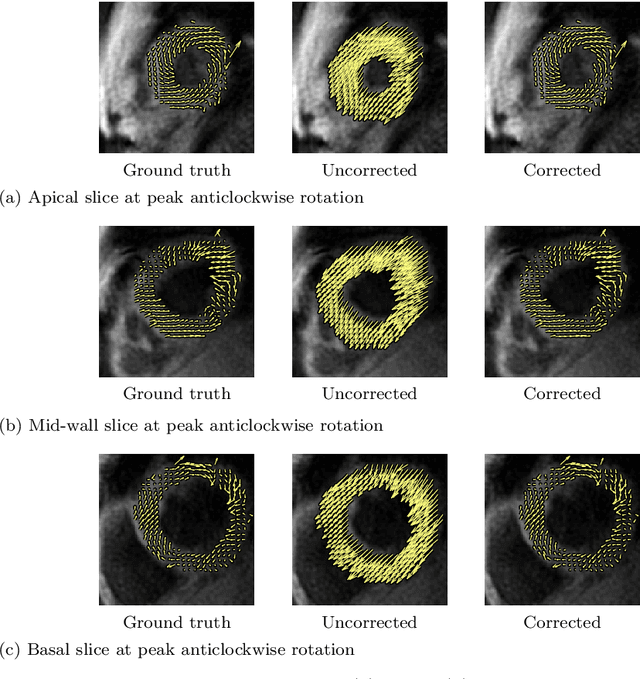
Abstract:Purpose: Advancements in MRI Tissue Phase Velocity Mapping (TPM) allow for the acquisition of higher quality velocity cardiac images providing better assessment of regional myocardial deformation for accurate disease diagnosis, pre-operative planning and post-operative patient surveillance. Translation of TPM velocities from the scanner's reference coordinate system to the regional cardiac coordinate system requires decoupling of translational motion and motion due to myocardial deformation. Despite existing techniques for respiratory motion compensation in TPM, there is still a remaining translational velocity component due to the global motion of the beating heart. To compensate for translational motion in cardiac TPM, we propose an image-processing method, which we have evaluated on synthetic data and applied on in vivo TPM data. Methods: Translational motion is estimated from a suitable region of velocities automatically defined in the left-ventricular volume. The region is generated by dilating the medial axis of myocardial masks in each slice and the translational velocity is estimated by integration in this region. The method was evaluated on synthetic data and in vivo data corrupted with a translational velocity component (200% of the maximum measured velocity). Accuracy and robustness were examined and the method was applied on 10 in vivo datasets. Results: The results from synthetic and in vivo corrupted data show excellent performance with an estimation error less than 0.3% and high robustness in both cases. The effectiveness of the method is confirmed with visual observation of results from the 10 datasets. Conclusion: The proposed method is accurate and suitable for translational motion correction of the left ventricular velocity fields. The current method for translational motion compensation could be applied to any annular contracting (tissue) structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge