David Firmin
HDL: Hybrid Deep Learning for the Synthesis of Myocardial Velocity Maps in Digital Twins for Cardiac Analysis
Mar 09, 2022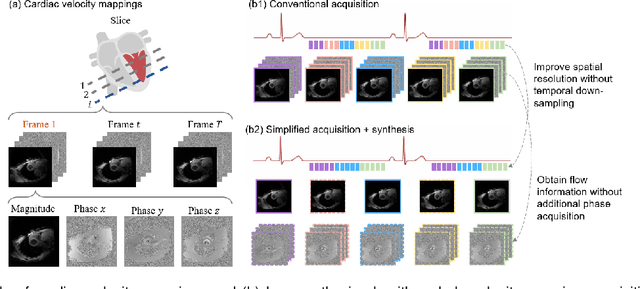
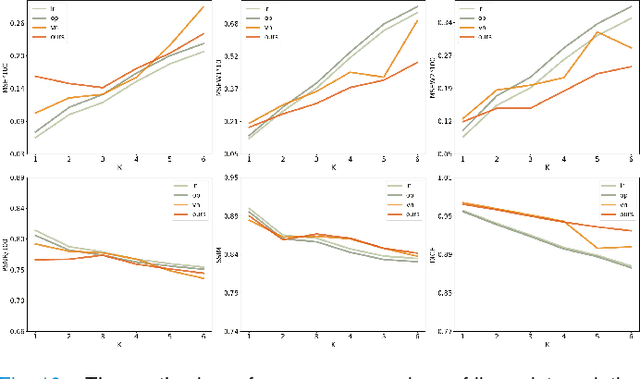
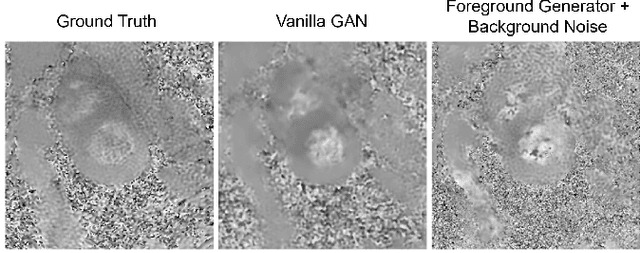
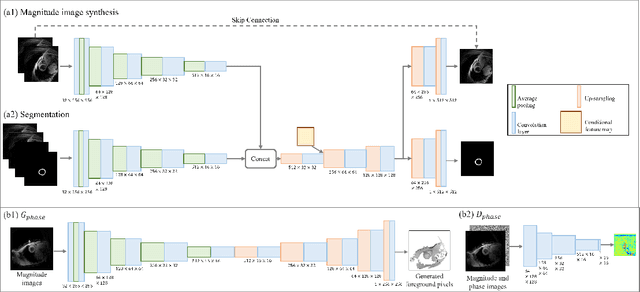
Abstract:Synthetic digital twins based on medical data accelerate the acquisition, labelling and decision making procedure in digital healthcare. A core part of digital healthcare twins is model-based data synthesis, which permits the generation of realistic medical signals without requiring to cope with the modelling complexity of anatomical and biochemical phenomena producing them in reality. Unfortunately, algorithms for cardiac data synthesis have been so far scarcely studied in the literature. An important imaging modality in the cardiac examination is three-directional CINE multi-slice myocardial velocity mapping (3Dir MVM), which provides a quantitative assessment of cardiac motion in three orthogonal directions of the left ventricle. The long acquisition time and complex acquisition produce make it more urgent to produce synthetic digital twins of this imaging modality. In this study, we propose a hybrid deep learning (HDL) network, especially for synthetic 3Dir MVM data. Our algorithm is featured by a hybrid UNet and a Generative Adversarial Network with a foreground-background generation scheme. The experimental results show that from temporally down-sampled magnitude CINE images (six times), our proposed algorithm can still successfully synthesise high temporal resolution 3Dir MVM CMR data (PSNR=42.32) with precise left ventricle segmentation (DICE=0.92). These performance scores indicate that our proposed HDL algorithm can be implemented in real-world digital twins for myocardial velocity mapping data simulation. To the best of our knowledge, this work is the first one in the literature investigating digital twins of the 3Dir MVM CMR, which has shown great potential for improving the efficiency of clinical studies via synthesised cardiac data.
AI-based Reconstruction for Fast MRI -- A Systematic Review and Meta-analysis
Dec 23, 2021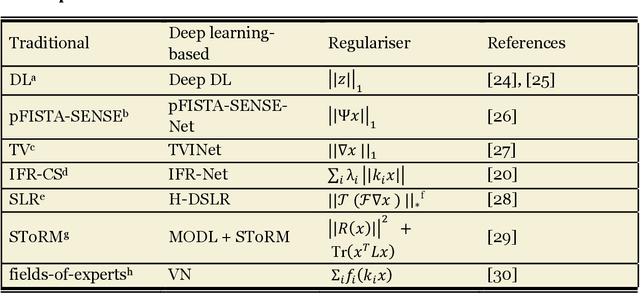
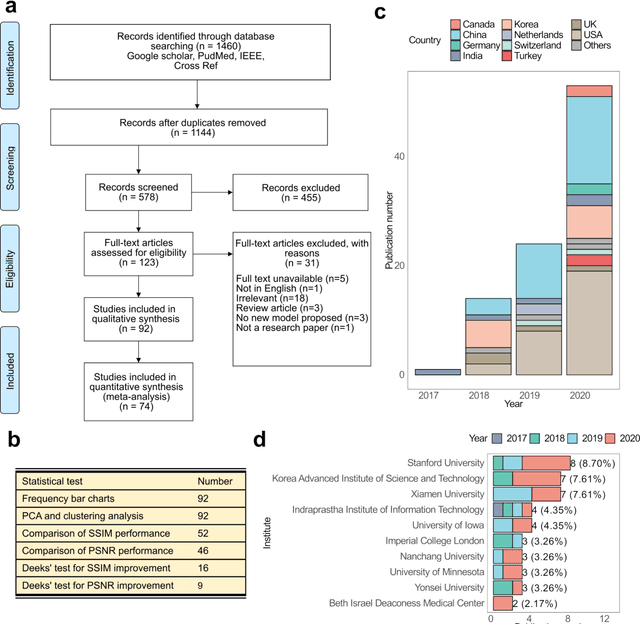
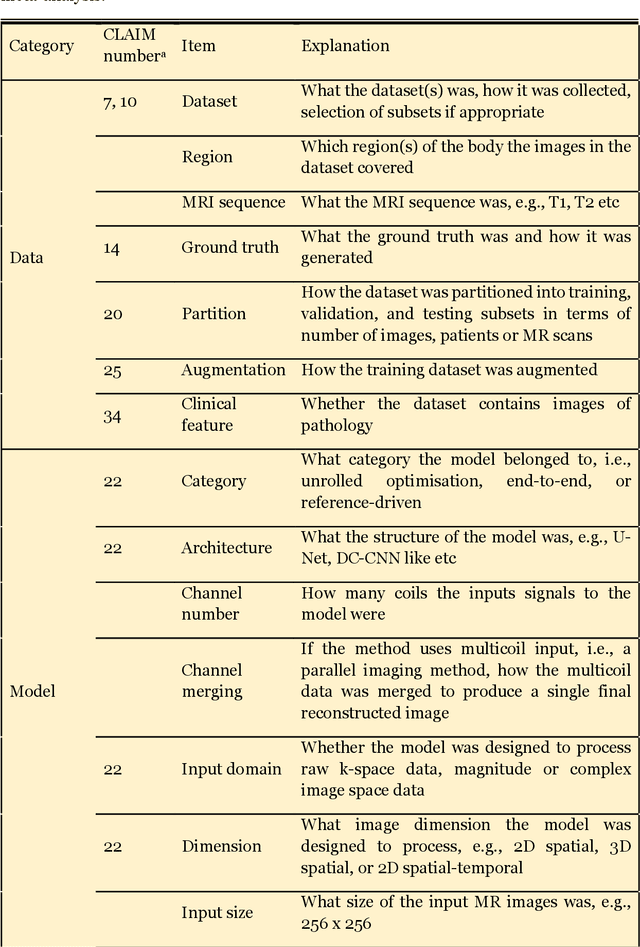
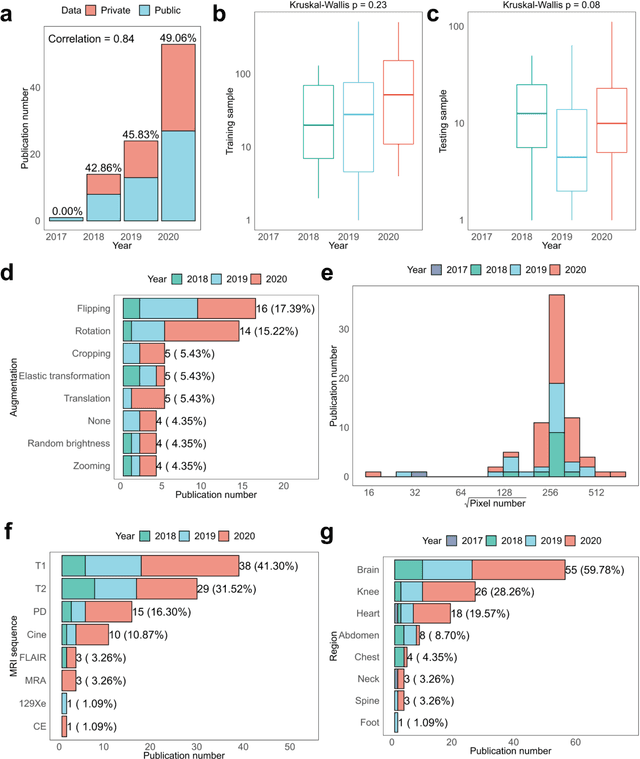
Abstract:Compressed sensing (CS) has been playing a key role in accelerating the magnetic resonance imaging (MRI) acquisition process. With the resurgence of artificial intelligence, deep neural networks and CS algorithms are being integrated to redefine the state of the art of fast MRI. The past several years have witnessed substantial growth in the complexity, diversity, and performance of deep learning-based CS techniques that are dedicated to fast MRI. In this meta-analysis, we systematically review the deep learning-based CS techniques for fast MRI, describe key model designs, highlight breakthroughs, and discuss promising directions. We have also introduced a comprehensive analysis framework and a classification system to assess the pivotal role of deep learning in CS-based acceleration for MRI.
Synthetic Velocity Mapping Cardiac MRI Coupled with Automated Left Ventricle Segmentation
Oct 04, 2021

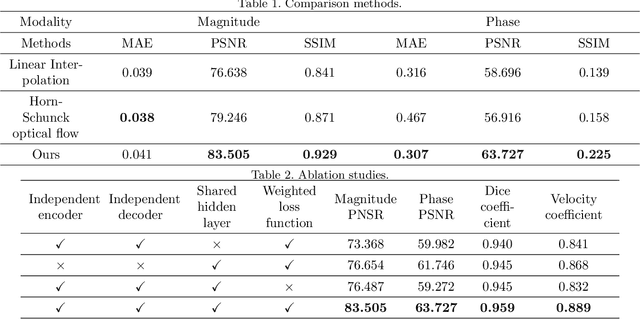

Abstract:Temporal patterns of cardiac motion provide important information for cardiac disease diagnosis. This pattern could be obtained by three-directional CINE multi-slice left ventricular myocardial velocity mapping (3Dir MVM), which is a cardiac MR technique providing magnitude and phase information of the myocardial motion simultaneously. However, long acquisition time limits the usage of this technique by causing breathing artifacts, while shortening the time causes low temporal resolution and may provide an inaccurate assessment of cardiac motion. In this study, we proposed a frame synthesis algorithm to increase the temporal resolution of 3Dir MVM data. Our algorithm is featured by 1) three attention-based encoders which accept magnitude images, phase images, and myocardium segmentation masks respectively as inputs; 2) three decoders that output the interpolated frames and corresponding myocardium segmentation results; and 3) loss functions highlighting myocardium pixels. Our algorithm can not only increase the temporal resolution 3Dir MVMs, but can also generates the myocardium segmentation results at the same time.
Adaptive Hierarchical Dual Consistency for Semi-Supervised Left Atrium Segmentation on Cross-Domain Data
Sep 20, 2021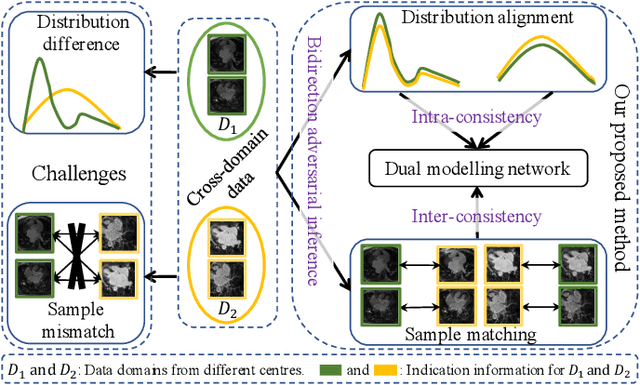
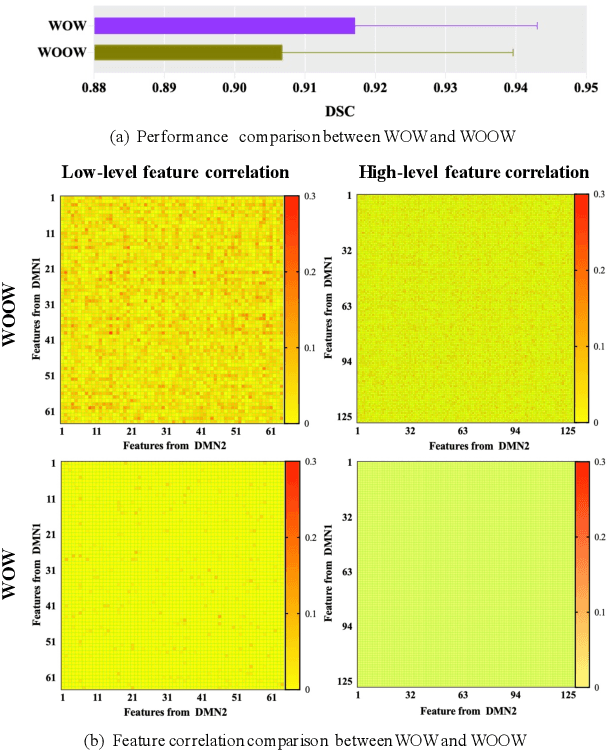
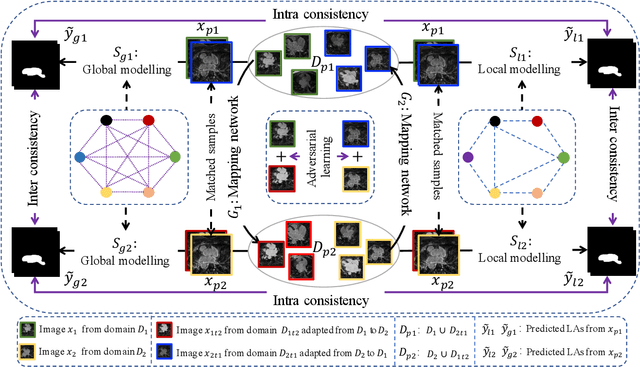
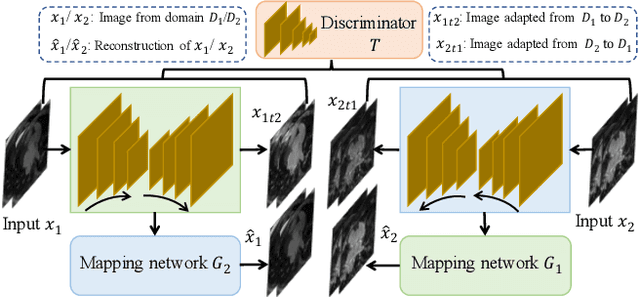
Abstract:Semi-supervised learning provides great significance in left atrium (LA) segmentation model learning with insufficient labelled data. Generalising semi-supervised learning to cross-domain data is of high importance to further improve model robustness. However, the widely existing distribution difference and sample mismatch between different data domains hinder the generalisation of semi-supervised learning. In this study, we alleviate these problems by proposing an Adaptive Hierarchical Dual Consistency (AHDC) for the semi-supervised LA segmentation on cross-domain data. The AHDC mainly consists of a Bidirectional Adversarial Inference module (BAI) and a Hierarchical Dual Consistency learning module (HDC). The BAI overcomes the difference of distributions and the sample mismatch between two different domains. It mainly learns two mapping networks adversarially to obtain two matched domains through mutual adaptation. The HDC investigates a hierarchical dual learning paradigm for cross-domain semi-supervised segmentation based on the obtained matched domains. It mainly builds two dual-modelling networks for mining the complementary information in both intra-domain and inter-domain. For the intra-domain learning, a consistency constraint is applied to the dual-modelling targets to exploit the complementary modelling information. For the inter-domain learning, a consistency constraint is applied to the LAs modelled by two dual-modelling networks to exploit the complementary knowledge among different data domains. We demonstrated the performance of our proposed AHDC on four 3D late gadolinium enhancement cardiac MR (LGE-CMR) datasets from different centres and a 3D CT dataset. Compared to other state-of-the-art methods, our proposed AHDC achieved higher segmentation accuracy, which indicated its capability in the cross-domain semi-supervised LA segmentation.
Recent Advances in Fibrosis and Scar Segmentation from Cardiac MRI: A State-of-the-Art Review and Future Perspectives
Jun 28, 2021



Abstract:Segmentation of cardiac fibrosis and scar are essential for clinical diagnosis and can provide invaluable guidance for the treatment of cardiac diseases. Late Gadolinium enhancement (LGE) cardiovascular magnetic resonance (CMR) has been successful for its efficacy in guiding the clinical diagnosis and treatment reliably. For LGE CMR, many methods have demonstrated success in accurately segmenting scarring regions. Co-registration with other non-contrast-agent (non-CA) modalities, balanced steady-state free precession (bSSFP) and cine magnetic resonance imaging (MRI) for example, can further enhance the efficacy of automated segmentation of cardiac anatomies. Many conventional methods have been proposed to provide automated or semi-automated segmentation of scars. With the development of deep learning in recent years, we can also see more advanced methods that are more efficient in providing more accurate segmentations. This paper conducts a state-of-the-art review of conventional and current state-of-the-art approaches utilising different modalities for accurate cardiac fibrosis and scar segmentation.
JAS-GAN: Generative Adversarial Network Based Joint Atrium and Scar Segmentations on Unbalanced Atrial Targets
May 01, 2021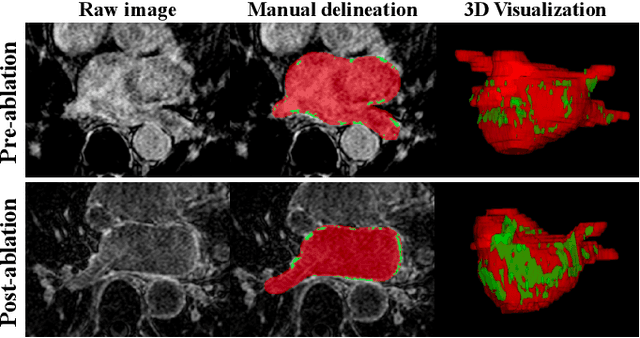
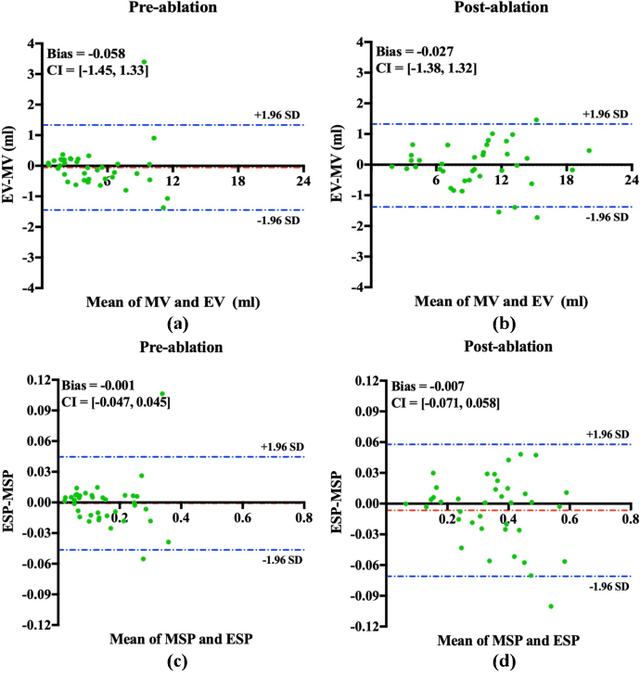
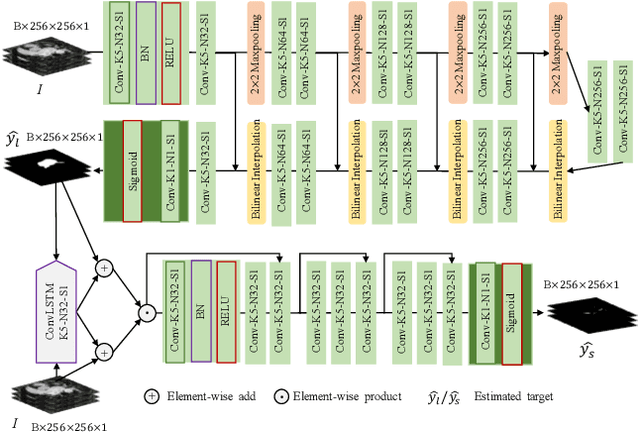
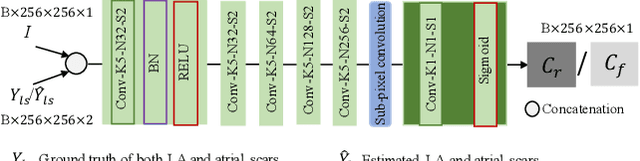
Abstract:Automated and accurate segmentations of left atrium (LA) and atrial scars from late gadolinium-enhanced cardiac magnetic resonance (LGE CMR) images are in high demand for quantifying atrial scars. The previous quantification of atrial scars relies on a two-phase segmentation for LA and atrial scars due to their large volume difference (unbalanced atrial targets). In this paper, we propose an inter-cascade generative adversarial network, namely JAS-GAN, to segment the unbalanced atrial targets from LGE CMR images automatically and accurately in an end-to-end way. Firstly, JAS-GAN investigates an adaptive attention cascade to automatically correlate the segmentation tasks of the unbalanced atrial targets. The adaptive attention cascade mainly models the inclusion relationship of the two unbalanced atrial targets, where the estimated LA acts as the attention map to adaptively focus on the small atrial scars roughly. Then, an adversarial regularization is applied to the segmentation tasks of the unbalanced atrial targets for making a consistent optimization. It mainly forces the estimated joint distribution of LA and atrial scars to match the real ones. We evaluated the performance of our JAS-GAN on a 3D LGE CMR dataset with 192 scans. Compared with the state-of-the-art methods, our proposed approach yielded better segmentation performance (Average Dice Similarity Coefficient (DSC) values of 0.946 and 0.821 for LA and atrial scars, respectively), which indicated the effectiveness of our proposed approach for segmenting unbalanced atrial targets.
Three-Dimensional Embedded Attentive RNN (3D-EAR) Segmentor for Left Ventricle Delineation from Myocardial Velocity Mapping
Apr 26, 2021
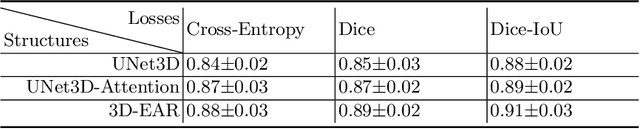
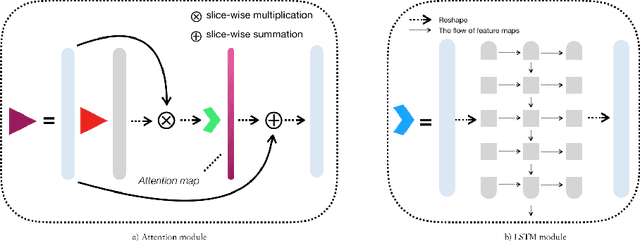
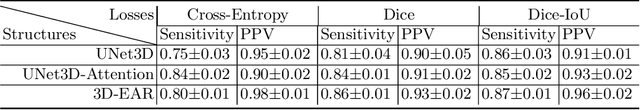
Abstract:Myocardial Velocity Mapping Cardiac MR (MVM-CMR) can be used to measure global and regional myocardial velocities with proved reproducibility. Accurate left ventricle delineation is a prerequisite for robust and reproducible myocardial velocity estimation. Conventional manual segmentation on this dataset can be time-consuming and subjective, and an effective fully automated delineation method is highly in demand. By leveraging recently proposed deep learning-based semantic segmentation approaches, in this study, we propose a novel fully automated framework incorporating a 3D-UNet backbone architecture with Embedded multichannel Attention mechanism and LSTM based Recurrent neural networks (RNN) for the MVM-CMR datasets (dubbed 3D-EAR segmentor). The proposed method also utilises the amalgamation of magnitude and phase images as input to realise an information fusion of this multichannel dataset and exploring the correlations of temporal frames via the embedded RNN. By comparing the baseline model of 3D-UNet and ablation studies with and without embedded attentive LSTM modules and various loss functions, we can demonstrate that the proposed model has outperformed the state-of-the-art baseline models with significant improvement.
Automated Multi-Channel Segmentation for the 4D Myocardial Velocity Mapping Cardiac MR
Dec 16, 2020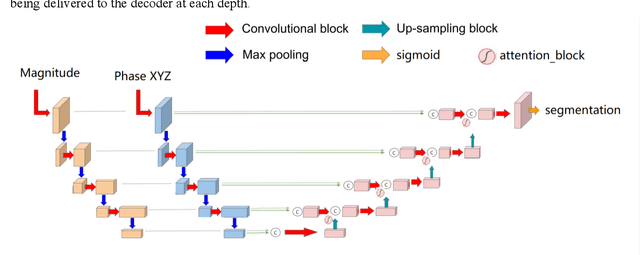
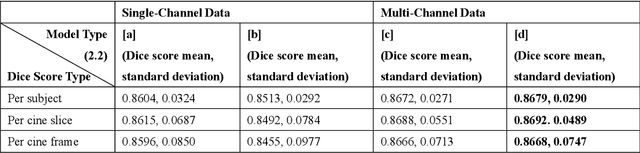

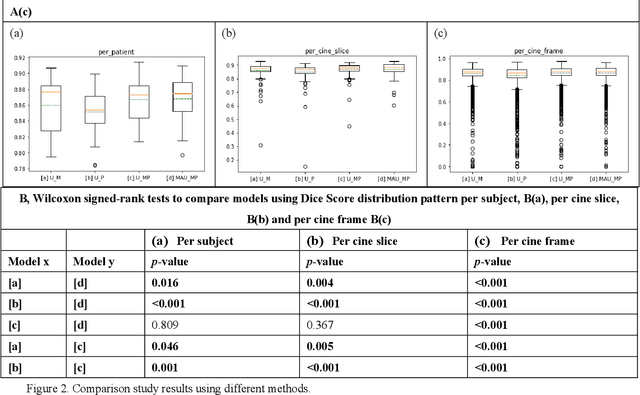
Abstract:Four-dimensional (4D) left ventricular myocardial velocity mapping (MVM) is a cardiac magnetic resonance (CMR) technique that allows assessment of cardiac motion in three orthogonal directions. Accurate and reproducible delineation of the myocardium is crucial for accurate analysis of peak systolic and diastolic myocardial velocities. In addition to the conventionally available magnitude CMR data, 4D MVM also acquires three velocity-encoded phase datasets which are used to generate velocity maps. These can be used to facilitate and improve myocardial delineation. Based on the success of deep learning in medical image processing, we propose a novel automated framework that improves the standard U-Net based methods on these CMR multi-channel data (magnitude and phase) by cross-channel fusion with attention module and shape information based post-processing to achieve accurate delineation of both epicardium and endocardium contours. To evaluate the results, we employ the widely used Dice scores and the quantification of myocardial longitudinal peak velocities. Our proposed network trained with multi-channel data shows enhanced performance compared to standard U-Net based networks trained with single-channel data. Based on the results, our method provides compelling evidence for the design and application for the multi-channel image analysis of the 4D MVM CMR data.
Simultaneous Left Atrium Anatomy and Scar Segmentations via Deep Learning in Multiview Information with Attention
Feb 02, 2020
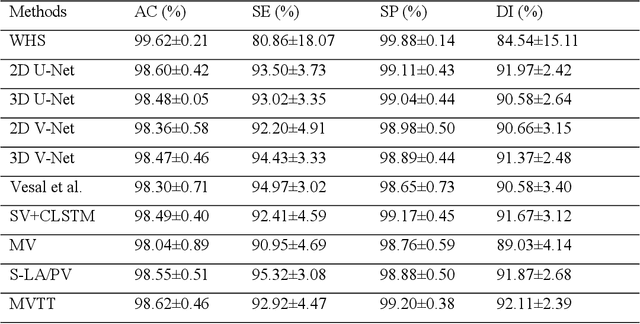

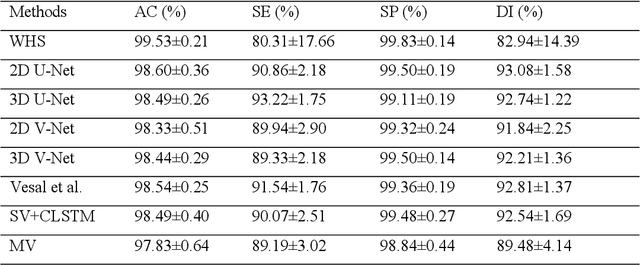
Abstract:Three-dimensional late gadolinium enhanced (LGE) cardiac MR (CMR) of left atrial scar in patients with atrial fibrillation (AF) has recently emerged as a promising technique to stratify patients, to guide ablation therapy and to predict treatment success. This requires a segmentation of the high intensity scar tissue and also a segmentation of the left atrium (LA) anatomy, the latter usually being derived from a separate bright-blood acquisition. Performing both segmentations automatically from a single 3D LGE CMR acquisition would eliminate the need for an additional acquisition and avoid subsequent registration issues. In this paper, we propose a joint segmentation method based on multiview two-task (MVTT) recursive attention model working directly on 3D LGE CMR images to segment the LA (and proximal pulmonary veins) and to delineate the scar on the same dataset. Using our MVTT recursive attention model, both the LA anatomy and scar can be segmented accurately (mean Dice score of 93% for the LA anatomy and 87% for the scar segmentations) and efficiently (~0.27 seconds to simultaneously segment the LA anatomy and scars directly from the 3D LGE CMR dataset with 60-68 2D slices). Compared to conventional unsupervised learning and other state-of-the-art deep learning based methods, the proposed MVTT model achieved excellent results, leading to an automatic generation of a patient-specific anatomical model combined with scar segmentation for patients in AF.
Discriminative Consistent Domain Generation for Semi-supervised Learning
Jul 24, 2019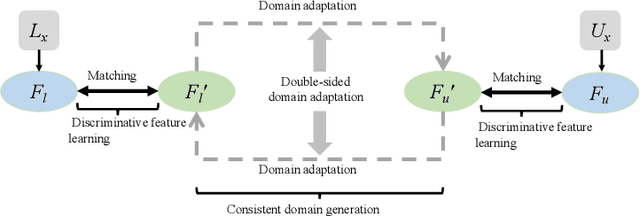
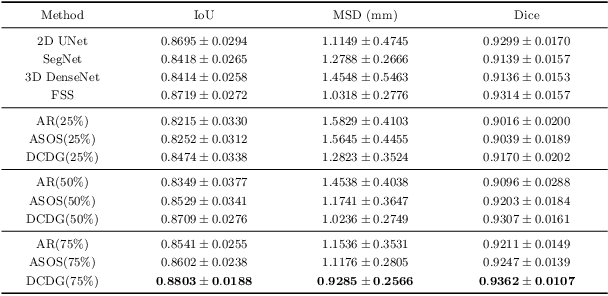
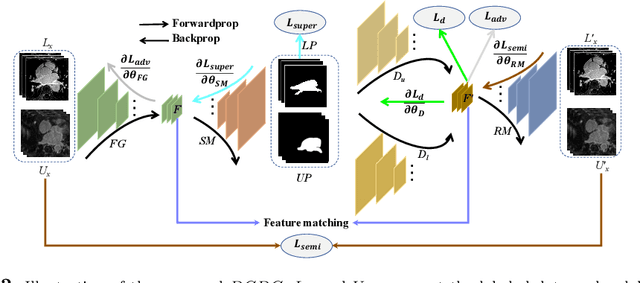
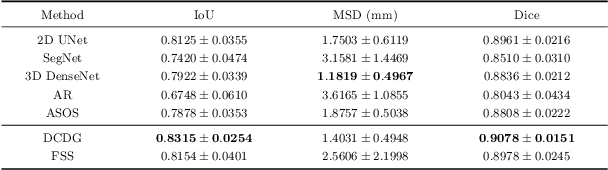
Abstract:Deep learning based task systems normally rely on a large amount of manually labeled training data, which is expensive to obtain and subject to operator variations. Moreover, it does not always hold that the manually labeled data and the unlabeled data are sitting in the same distribution. In this paper, we alleviate these problems by proposing a discriminative consistent domain generation (DCDG) approach to achieve a semi-supervised learning. The discriminative consistent domain is achieved by a double-sided domain adaptation. The double-sided domain adaptation aims to make a fusion of the feature spaces of labeled data and unlabeled data. In this way, we can fit the differences of various distributions between labeled data and unlabeled data. In order to keep the discriminativeness of generated consistent domain for the task learning, we apply an indirect learning for the double-sided domain adaptation. Based on the generated discriminative consistent domain, we can use the unlabeled data to learn the task model along with the labeled data via a consistent image generation. We demonstrate the performance of our proposed DCDG on the late gadolinium enhancement cardiac MRI (LGE-CMRI) images acquired from patients with atrial fibrillation in two clinical centers for the segmentation of the left atrium anatomy (LA) and proximal pulmonary veins (PVs). The experiments show that our semi-supervised approach achieves compelling segmentation results, which can prove the robustness of DCDG for the semi-supervised learning using the unlabeled data along with labeled data acquired from a single center or multicenter studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge