Habib Khan
Towards Automated Diagnosis of Inherited Arrhythmias: Combined Arrhythmia Classification Using Lead-Aware Spatial Attention Networks
Jan 12, 2026Abstract:Arrhythmogenic right ventricular cardiomyopathy (ARVC) and long QT syndrome (LQTS) are inherited arrhythmia syndromes associated with sudden cardiac death. Deep learning shows promise for ECG interpretation, but multi-class inherited arrhythmia classification with clinically grounded interpretability remains underdeveloped. Our objective was to develop and validate a lead-aware deep learning framework for multi-class (ARVC vs LQTS vs control) and binary inherited arrhythmia classification, and to determine optimal strategies for integrating ECG foundation models within arrhythmia screening tools. We assembled a 13-center Canadian cohort (645 patients; 1,344 ECGs). We evaluated four ECG foundation models using three transfer learning approaches: linear probing, fine-tuning, and combined strategies. We developed lead-aware spatial attention networks (LASAN) and assessed integration strategies combining LASAN with foundation models. Performance was compared against the established foundation model baselines. Lead-group masking quantified disease-specific lead dependence. Fine-tuning outperformed linear probing and combined strategies across all foundation models (mean macro-AUROC 0.904 vs 0.825). The best lead-aware integrations achieved near-ceiling performance (HuBERT-ECG hybrid: macro-AUROC 0.990; ARVC vs control AUROC 0.999; LQTS vs control AUROC 0.994). Lead masking demonstrated physiologic plausibility: V1-V3 were most critical for ARVC detection (4.54% AUROC reduction), while lateral leads were preferentially important for LQTS (2.60% drop). Lead-aware architectures achieved state-of-the-art performance for inherited arrhythmia classification, outperforming all existing published models on both binary and multi-class tasks while demonstrating clinically aligned lead dependence. These findings support potential utility for automated ECG screening pending validation.
JAS-GAN: Generative Adversarial Network Based Joint Atrium and Scar Segmentations on Unbalanced Atrial Targets
May 01, 2021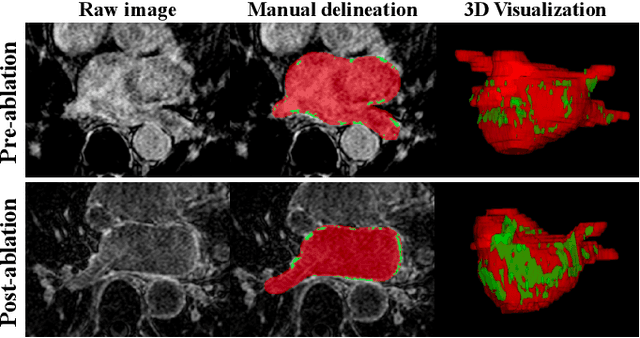
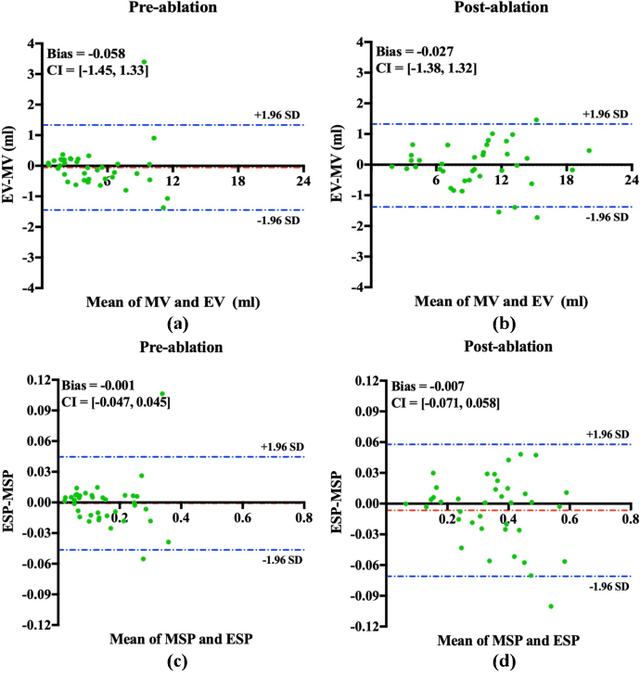
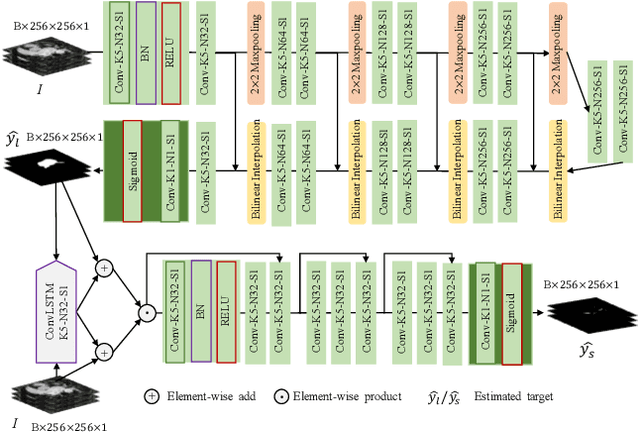
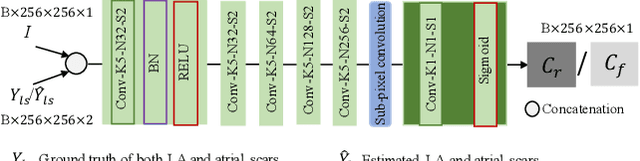
Abstract:Automated and accurate segmentations of left atrium (LA) and atrial scars from late gadolinium-enhanced cardiac magnetic resonance (LGE CMR) images are in high demand for quantifying atrial scars. The previous quantification of atrial scars relies on a two-phase segmentation for LA and atrial scars due to their large volume difference (unbalanced atrial targets). In this paper, we propose an inter-cascade generative adversarial network, namely JAS-GAN, to segment the unbalanced atrial targets from LGE CMR images automatically and accurately in an end-to-end way. Firstly, JAS-GAN investigates an adaptive attention cascade to automatically correlate the segmentation tasks of the unbalanced atrial targets. The adaptive attention cascade mainly models the inclusion relationship of the two unbalanced atrial targets, where the estimated LA acts as the attention map to adaptively focus on the small atrial scars roughly. Then, an adversarial regularization is applied to the segmentation tasks of the unbalanced atrial targets for making a consistent optimization. It mainly forces the estimated joint distribution of LA and atrial scars to match the real ones. We evaluated the performance of our JAS-GAN on a 3D LGE CMR dataset with 192 scans. Compared with the state-of-the-art methods, our proposed approach yielded better segmentation performance (Average Dice Similarity Coefficient (DSC) values of 0.946 and 0.821 for LA and atrial scars, respectively), which indicated the effectiveness of our proposed approach for segmenting unbalanced atrial targets.
Fully Automatic Segmentation and Objective Assessment of Atrial Scars for Longstanding Persistent Atrial Fibrillation Patients Using Late Gadolinium-Enhanced MRI
May 26, 2017



Abstract:Purpose: Atrial fibrillation (AF) is the most common cardiac arrhythmia and is correlated with increased morbidity and mortality. It is associated with atrial fibrosis, which may be assessed non-invasively using late gadolinium-enhanced (LGE) magnetic resonance imaging (MRI) where scar tissue is visualised as a region of signal enhancement. In this study, we proposed a novel fully automatic pipeline to achieve an accurate and objective atrial scarring segmentation and assessment of LGE MRI scans for the AF patients. Methods: Our fully automatic pipeline uniquely combined: (1) a multi-atlas based whole heart segmentation (MA-WHS) to determine the cardiac anatomy from an MRI Roadmap acquisition which is then mapped to LGE MRI, and (2) a super-pixel and supervised learning based approach to delineate the distribution and extent of atrial scarring in LGE MRI. Results: Both our MA-WHS and atrial scarring segmentation showed accurate delineations of cardiac anatomy (mean Dice = 89%) and atrial scarring (mean Dice =79%) respectively compared to the established ground truth from manual segmentation. Compared with previously studied methods with manual interventions, our innovative pipeline demonstrated comparable results, but was computed fully automatically. Conclusion: The proposed segmentation methods allow LGE MRI to be used as an objective assessment tool for localisation, visualisation and quantification of atrial scarring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge