Jascha Achterberg
Algorithm-hardware co-design of neuromorphic networks with dual memory pathways
Dec 11, 2025Abstract:Spiking neural networks excel at event-driven sensing. Yet, maintaining task-relevant context over long timescales both algorithmically and in hardware, while respecting both tight energy and memory budgets, remains a core challenge in the field. We address this challenge through novel algorithm-hardware co-design effort. At the algorithm level, inspired by the cortical fast-slow organization in the brain, we introduce a neural network with an explicit slow memory pathway that, combined with fast spiking activity, enables a dual memory pathway (DMP) architecture in which each layer maintains a compact low-dimensional state that summarizes recent activity and modulates spiking dynamics. This explicit memory stabilizes learning while preserving event-driven sparsity, achieving competitive accuracy on long-sequence benchmarks with 40-60% fewer parameters than equivalent state-of-the-art spiking neural networks. At the hardware level, we introduce a near-memory-compute architecture that fully leverages the advantages of the DMP architecture by retaining its compact shared state while optimizing dataflow, across heterogeneous sparse-spike and dense-memory pathways. We show experimental results that demonstrate more than a 4x increase in throughput and over a 5x improvement in energy efficiency compared with state-of-the-art implementations. Together, these contributions demonstrate that biological principles can guide functional abstractions that are both algorithmically effective and hardware-efficient, establishing a scalable co-design paradigm for real-time neuromorphic computation and learning.
Exploiting heterogeneous delays for efficient computation in low-bit neural networks
Oct 31, 2025Abstract:Neural networks rely on learning synaptic weights. However, this overlooks other neural parameters that can also be learned and may be utilized by the brain. One such parameter is the delay: the brain exhibits complex temporal dynamics with heterogeneous delays, where signals are transmitted asynchronously between neurons. It has been theorized that this delay heterogeneity, rather than a cost to be minimized, can be exploited in embodied contexts where task-relevant information naturally sits contextually in the time domain. We test this hypothesis by training spiking neural networks to modify not only their weights but also their delays at different levels of precision. We find that delay heterogeneity enables state-of-the-art performance on temporally complex neuromorphic problems and can be achieved even when weights are extremely imprecise (1.58-bit ternary precision: just positive, negative, or absent). By enabling high performance with extremely low-precision weights, delay heterogeneity allows memory-efficient solutions that maintain state-of-the-art accuracy even when weights are compressed over an order of magnitude more aggressively than typically studied weight-only networks. We show how delays and time-constants adaptively trade-off, and reveal through ablation that task performance depends on task-appropriate delay distributions, with temporally-complex tasks requiring longer delays. Our results suggest temporal heterogeneity is an important principle for efficient computation, particularly when task-relevant information is temporal - as in the physical world - with implications for embodied intelligent systems and neuromorphic hardware.
Dynamical similarity analysis uniquely captures how computations develop in RNNs
Oct 31, 2024



Abstract:Methods for analyzing representations in neural systems are increasingly popular tools in neuroscience and mechanistic interpretability. Measures comparing neural activations across conditions, architectures, and species give scalable ways to understand information transformation within different neural networks. However, recent findings show that some metrics respond to spurious signals, leading to misleading results. Establishing benchmark test cases is thus essential for identifying the most reliable metric and potential improvements. We propose that compositional learning in recurrent neural networks (RNNs) can provide a test case for dynamical representation alignment metrics. Implementing this case allows us to evaluate if metrics can identify representations that develop throughout learning and determine if representations identified by metrics reflect the network's actual computations. Building both attractor and RNN based test cases, we show that the recently proposed Dynamical Similarity Analysis (DSA) is more noise robust and reliably identifies behaviorally relevant representations compared to prior metrics (Procrustes, CKA). We also demonstrate how such test cases can extend beyond metric evaluation to study new architectures. Specifically, testing DSA in modern (Mamba) state space models suggests that these models, unlike RNNs, may not require changes in recurrent dynamics due to their expressive hidden states. Overall, we develop test cases that showcase how DSA's enhanced ability to detect dynamical motifs makes it highly effective for identifying ongoing computations in RNNs and revealing how networks learn tasks.
Accelerated AI Inference via Dynamic Execution Methods
Oct 30, 2024


Abstract:In this paper, we focus on Dynamic Execution techniques that optimize the computation flow based on input. This aims to identify simpler problems that can be solved using fewer resources, similar to human cognition. The techniques discussed include early exit from deep networks, speculative sampling for language models, and adaptive steps for diffusion models. Experimental results demonstrate that these dynamic approaches can significantly improve latency and throughput without compromising quality. When combined with model-based optimizations, such as quantization, dynamic execution provides a powerful multi-pronged strategy to optimize AI inference. Generative AI requires a large amount of compute resources. This is expected to grow, and demand for resources in data centers through to the edge is expected to continue to increase at high rates. We take advantage of existing research and provide additional innovations for some generative optimizations. In the case of LLMs, we provide more efficient sampling methods that depend on the complexity of the data. In the case of diffusion model generation, we provide a new method that also leverages the difficulty of the input prompt to predict an optimal early stopping point. Therefore, dynamic execution methods are relevant because they add another dimension of performance optimizations. Performance is critical from a competitive point of view, but increasing capacity can result in significant power savings and cost savings. We have provided several integrations of these techniques into several Intel performance libraries and Huggingface Optimum. These integrations will make them easier to use and increase the adoption of these techniques.
Spatial embedding promotes a specific form of modularity with low entropy and heterogeneous spectral dynamics
Sep 26, 2024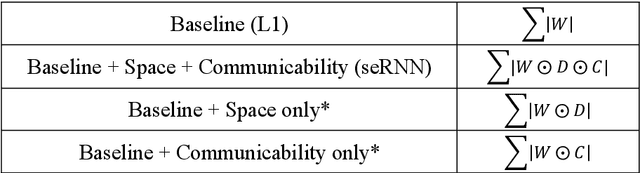
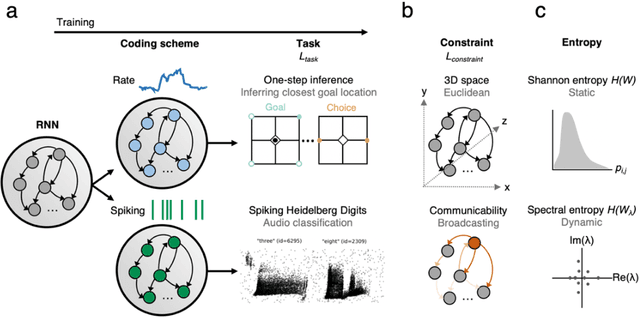
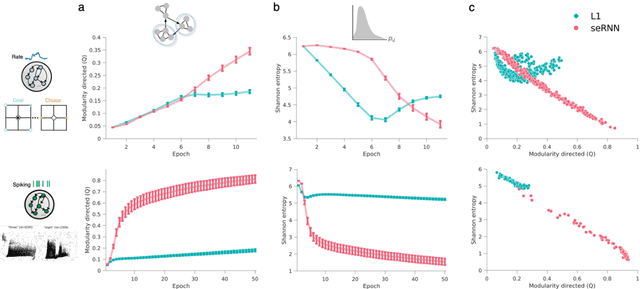
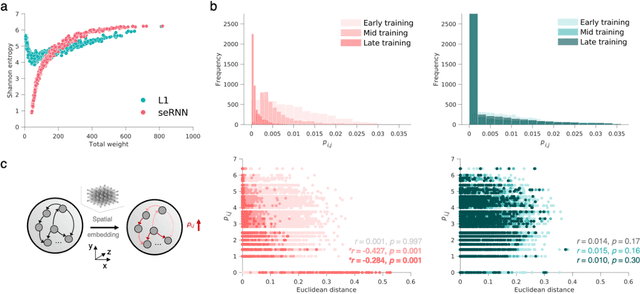
Abstract:Understanding how biological constraints shape neural computation is a central goal of computational neuroscience. Spatially embedded recurrent neural networks provide a promising avenue to study how modelled constraints shape the combined structural and functional organisation of networks over learning. Prior work has shown that spatially embedded systems like this can combine structure and function into single artificial models during learning. But it remains unclear precisely how, in general, structural constraints bound the range of attainable configurations. In this work, we show that it is possible to study these restrictions through entropic measures of the neural weights and eigenspectrum, across both rate and spiking neural networks. Spatial embedding, in contrast to baseline models, leads to networks with a highly specific low entropy modularity where connectivity is readily interpretable given the known spatial and communication constraints acting on them. Crucially, these networks also demonstrate systematically modulated spectral dynamics, revealing how they exploit heterogeneity in their function to overcome the constraints imposed on their structure. This work deepens our understanding of constrained learning in neural networks, across coding schemes and tasks, where solutions to simultaneous structural and functional objectives must be accomplished in tandem.
Multilevel Interpretability Of Artificial Neural Networks: Leveraging Framework And Methods From Neuroscience
Aug 26, 2024
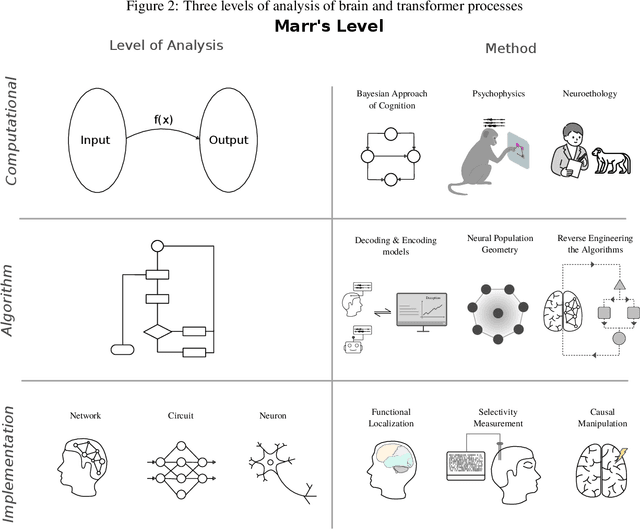
Abstract:As deep learning systems are scaled up to many billions of parameters, relating their internal structure to external behaviors becomes very challenging. Although daunting, this problem is not new: Neuroscientists and cognitive scientists have accumulated decades of experience analyzing a particularly complex system - the brain. In this work, we argue that interpreting both biological and artificial neural systems requires analyzing those systems at multiple levels of analysis, with different analytic tools for each level. We first lay out a joint grand challenge among scientists who study the brain and who study artificial neural networks: understanding how distributed neural mechanisms give rise to complex cognition and behavior. We then present a series of analytical tools that can be used to analyze biological and artificial neural systems, organizing those tools according to Marr's three levels of analysis: computation/behavior, algorithm/representation, and implementation. Overall, the multilevel interpretability framework provides a principled way to tackle neural system complexity; links structure, computation, and behavior; clarifies assumptions and research priorities at each level; and paves the way toward a unified effort for understanding intelligent systems, may they be biological or artificial.
Getting aligned on representational alignment
Nov 02, 2023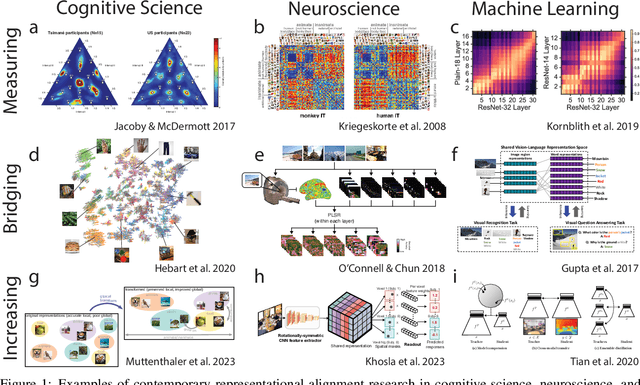
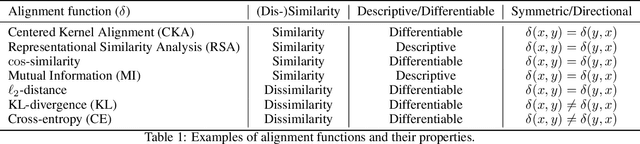
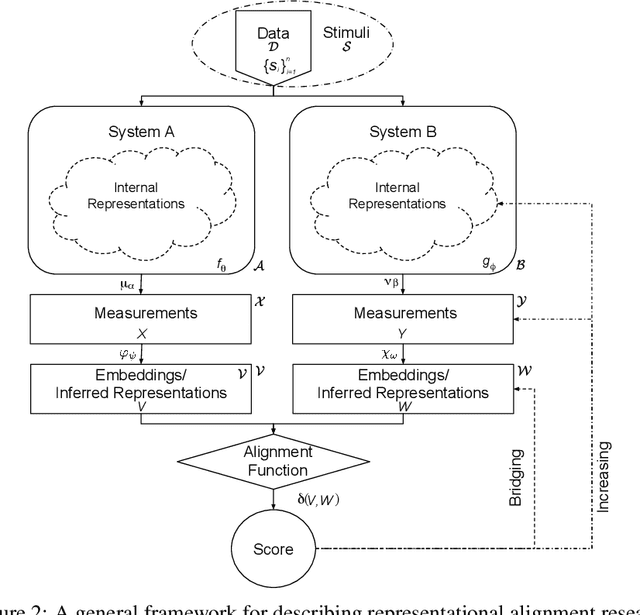
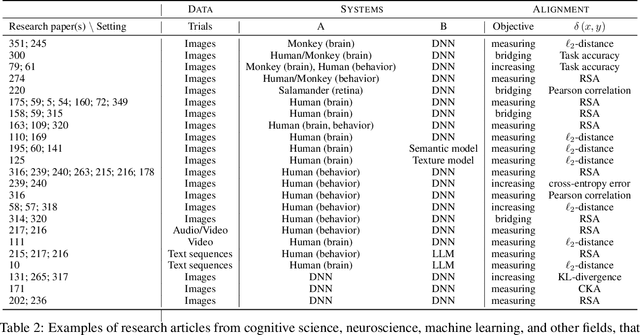
Abstract:Biological and artificial information processing systems form representations that they can use to categorize, reason, plan, navigate, and make decisions. How can we measure the extent to which the representations formed by these diverse systems agree? Do similarities in representations then translate into similar behavior? How can a system's representations be modified to better match those of another system? These questions pertaining to the study of representational alignment are at the heart of some of the most active research areas in cognitive science, neuroscience, and machine learning. For example, cognitive scientists measure the representational alignment of multiple individuals to identify shared cognitive priors, neuroscientists align fMRI responses from multiple individuals into a shared representational space for group-level analyses, and ML researchers distill knowledge from teacher models into student models by increasing their alignment. Unfortunately, there is limited knowledge transfer between research communities interested in representational alignment, so progress in one field often ends up being rediscovered independently in another. Thus, greater cross-field communication would be advantageous. To improve communication between these fields, we propose a unifying framework that can serve as a common language between researchers studying representational alignment. We survey the literature from all three fields and demonstrate how prior work fits into this framework. Finally, we lay out open problems in representational alignment where progress can benefit all three of these fields. We hope that our work can catalyze cross-disciplinary collaboration and accelerate progress for all communities studying and developing information processing systems. We note that this is a working paper and encourage readers to reach out with their suggestions for future revisions.
Brain-inspired learning in artificial neural networks: a review
May 18, 2023


Abstract:Artificial neural networks (ANNs) have emerged as an essential tool in machine learning, achieving remarkable success across diverse domains, including image and speech generation, game playing, and robotics. However, there exist fundamental differences between ANNs' operating mechanisms and those of the biological brain, particularly concerning learning processes. This paper presents a comprehensive review of current brain-inspired learning representations in artificial neural networks. We investigate the integration of more biologically plausible mechanisms, such as synaptic plasticity, to enhance these networks' capabilities. Moreover, we delve into the potential advantages and challenges accompanying this approach. Ultimately, we pinpoint promising avenues for future research in this rapidly advancing field, which could bring us closer to understanding the essence of intelligence.
Building artificial neural circuits for domain-general cognition: a primer on brain-inspired systems-level architecture
Mar 21, 2023
Abstract:There is a concerted effort to build domain-general artificial intelligence in the form of universal neural network models with sufficient computational flexibility to solve a wide variety of cognitive tasks but without requiring fine-tuning on individual problem spaces and domains. To do this, models need appropriate priors and inductive biases, such that trained models can generalise to out-of-distribution examples and new problem sets. Here we provide an overview of the hallmarks endowing biological neural networks with the functionality needed for flexible cognition, in order to establish which features might also be important to achieve similar functionality in artificial systems. We specifically discuss the role of system-level distribution of network communication and recurrence, in addition to the role of short-term topological changes for efficient local computation. As machine learning models become more complex, these principles may provide valuable directions in an otherwise vast space of possible architectures. In addition, testing these inductive biases within artificial systems may help us to understand the biological principles underlying domain-general cognition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge