John Duncan
From Pre- to Intra-operative MRI: Predicting Brain Shift in Temporal Lobe Resection for Epilepsy Surgery
Feb 03, 2026Abstract:Introduction: In neurosurgery, image-guided Neurosurgery Systems (IGNS) highly rely on preoperative brain magnetic resonance images (MRI) to assist surgeons in locating surgical targets and determining surgical paths. However, brain shift invalidates the preoperative MRI after dural opening. Updated intraoperative brain MRI with brain shift compensation is crucial for enhancing the precision of neuronavigation systems and ensuring the optimal outcome of surgical interventions. Methodology: We propose NeuralShift, a U-Net-based model that predicts brain shift entirely from pre-operative MRI for patients undergoing temporal lobe resection. We evaluated our results using Target Registration Errors (TREs) computed on anatomical landmarks located on the resection side and along the midline, and DICE scores comparing predicted intraoperative masks with masks derived from intraoperative MRI. Results: Our experimental results show that our model can predict the global deformation of the brain (DICE of 0.97) with accurate local displacements (achieve landmark TRE as low as 1.12 mm), compensating for large brain shifts during temporal lobe removal neurosurgery. Conclusion: Our proposed model is capable of predicting the global deformation of the brain during temporal lobe resection using only preoperative images, providing potential opportunities to the surgical team to increase safety and efficiency of neurosurgery and better outcomes to patients. Our contributions will be publicly available after acceptance in https://github.com/SurgicalDataScienceKCL/NeuralShift.
Building artificial neural circuits for domain-general cognition: a primer on brain-inspired systems-level architecture
Mar 21, 2023
Abstract:There is a concerted effort to build domain-general artificial intelligence in the form of universal neural network models with sufficient computational flexibility to solve a wide variety of cognitive tasks but without requiring fine-tuning on individual problem spaces and domains. To do this, models need appropriate priors and inductive biases, such that trained models can generalise to out-of-distribution examples and new problem sets. Here we provide an overview of the hallmarks endowing biological neural networks with the functionality needed for flexible cognition, in order to establish which features might also be important to achieve similar functionality in artificial systems. We specifically discuss the role of system-level distribution of network communication and recurrence, in addition to the role of short-term topological changes for efficient local computation. As machine learning models become more complex, these principles may provide valuable directions in an otherwise vast space of possible architectures. In addition, testing these inductive biases within artificial systems may help us to understand the biological principles underlying domain-general cognition.
Using convolution neural networks to learn enhanced fiber orientation distribution models from commercially available diffusion magnetic resonance imaging
Aug 12, 2020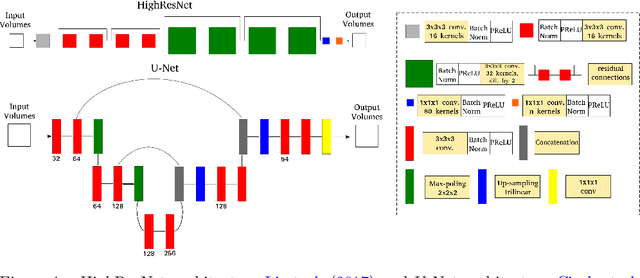
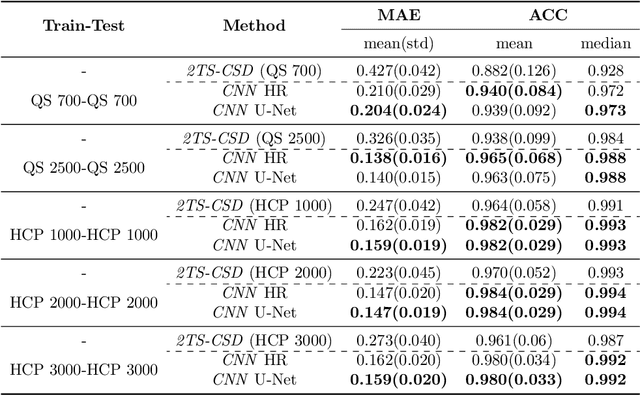
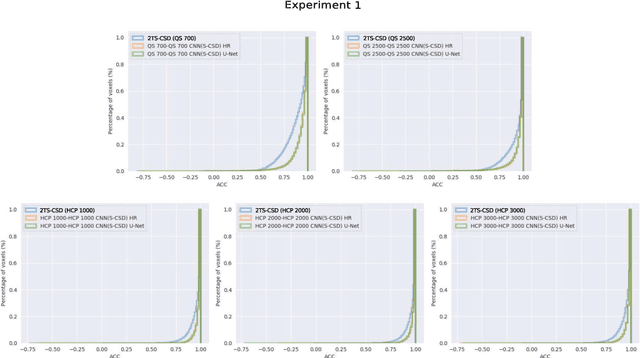
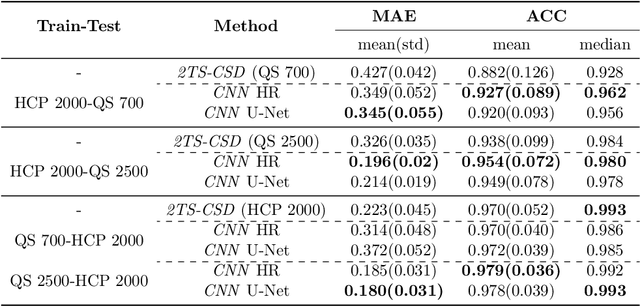
Abstract:Accurate local fiber orientation distribution (FOD) modeling based on diffusion magnetic resonance imaging (dMRI) capable of resolving complex fiber configurations benefit from specific acquisition protocols that impose a high number of gradient directions (b-vecs), a high maximum b-value (b-vals) and multiple b-values (multi-shell). However, acquisition time is limited in a clinical setting and commercial scanners may not provide robust state-of-the-art dMRI sequences. Therefore, dMRI is often acquired as single-shell (SS) (single b-value). Here, we learn improved FODs for commercially acquired dMRI. We evaluate the use of 3D convolutional neural networks (CNNs) to regress multi-shell FOS representations from single-shell representations, using the spherical harmonics basis obtained from constrained spherical deconvolution (CSD) to model FODs. We use U-Net and HighResNet 3D CNN architectures and data from the publicly available Human Connectome Dataset and a dataset acquired at National Hospital For Neurology and Neurosurgery Queen Square. We evaluate how well the CNN models can resolve local fiber orientation 1) when training and testing on datasets with same dMRI acquisition protocol; 2) when testing on dataset with a different dMRI acquisition protocol than used training the CNN models; and 3) when testing on datasets with a fewer number dMRI gradient directions than used training the CNN models. Our approach may enable robust CSD model estimation on dMRI acquisition protocols which are single shell and with a few gradient directions, reducing acquisition times, and thus, facilitating translation to time-limited clinical environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge