Using convolution neural networks to learn enhanced fiber orientation distribution models from commercially available diffusion magnetic resonance imaging
Paper and Code
Aug 12, 2020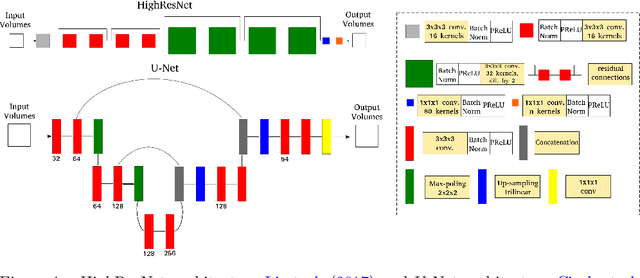
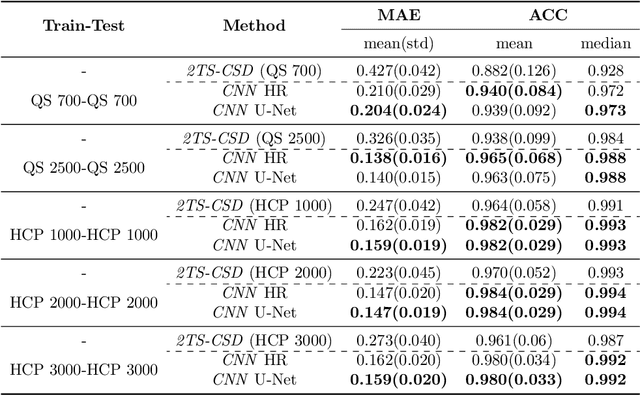
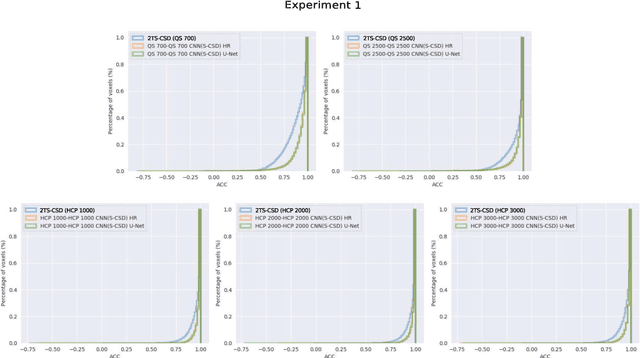
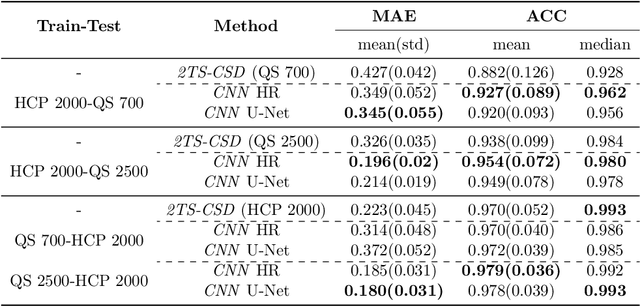
Accurate local fiber orientation distribution (FOD) modeling based on diffusion magnetic resonance imaging (dMRI) capable of resolving complex fiber configurations benefit from specific acquisition protocols that impose a high number of gradient directions (b-vecs), a high maximum b-value (b-vals) and multiple b-values (multi-shell). However, acquisition time is limited in a clinical setting and commercial scanners may not provide robust state-of-the-art dMRI sequences. Therefore, dMRI is often acquired as single-shell (SS) (single b-value). Here, we learn improved FODs for commercially acquired dMRI. We evaluate the use of 3D convolutional neural networks (CNNs) to regress multi-shell FOS representations from single-shell representations, using the spherical harmonics basis obtained from constrained spherical deconvolution (CSD) to model FODs. We use U-Net and HighResNet 3D CNN architectures and data from the publicly available Human Connectome Dataset and a dataset acquired at National Hospital For Neurology and Neurosurgery Queen Square. We evaluate how well the CNN models can resolve local fiber orientation 1) when training and testing on datasets with same dMRI acquisition protocol; 2) when testing on dataset with a different dMRI acquisition protocol than used training the CNN models; and 3) when testing on datasets with a fewer number dMRI gradient directions than used training the CNN models. Our approach may enable robust CSD model estimation on dMRI acquisition protocols which are single shell and with a few gradient directions, reducing acquisition times, and thus, facilitating translation to time-limited clinical environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge