Hang Hu
A Physics-driven GraphSAGE Method for Physical Process Simulations Described by Partial Differential Equations
Mar 13, 2024Abstract:Physics-informed neural networks (PINNs) have successfully addressed various computational physics problems based on partial differential equations (PDEs). However, while tackling issues related to irregularities like singularities and oscillations, trained solutions usually suffer low accuracy. In addition, most current works only offer the trained solution for predetermined input parameters. If any change occurs in input parameters, transfer learning or retraining is required, and traditional numerical techniques also need an independent simulation. In this work, a physics-driven GraphSAGE approach (PD-GraphSAGE) based on the Galerkin method and piecewise polynomial nodal basis functions is presented to solve computational problems governed by irregular PDEs and to develop parametric PDE surrogate models. This approach employs graph representations of physical domains, thereby reducing the demands for evaluated points due to local refinement. A distance-related edge feature and a feature mapping strategy are devised to help training and convergence for singularity and oscillation situations, respectively. The merits of the proposed method are demonstrated through a couple of cases. Moreover, the robust PDE surrogate model for heat conduction problems parameterized by the Gaussian random field source is successfully established, which not only provides the solution accurately but is several times faster than the finite element method in our experiments.
A Robust Deep Learning Method with Uncertainty Estimation for the Pathological Classification of Renal Cell Carcinoma based on CT Images
Nov 12, 2023



Abstract:Objectives To develop and validate a deep learning-based diagnostic model incorporating uncertainty estimation so as to facilitate radiologists in the preoperative differentiation of the pathological subtypes of renal cell carcinoma (RCC) based on CT images. Methods Data from 668 consecutive patients, pathologically proven RCC, were retrospectively collected from Center 1. By using five-fold cross-validation, a deep learning model incorporating uncertainty estimation was developed to classify RCC subtypes into clear cell RCC (ccRCC), papillary RCC (pRCC), and chromophobe RCC (chRCC). An external validation set of 78 patients from Center 2 further evaluated the model's performance. Results In the five-fold cross-validation, the model's area under the receiver operating characteristic curve (AUC) for the classification of ccRCC, pRCC, and chRCC was 0.868 (95% CI: 0.826-0.923), 0.846 (95% CI: 0.812-0.886), and 0.839 (95% CI: 0.802-0.88), respectively. In the external validation set, the AUCs were 0.856 (95% CI: 0.838-0.882), 0.787 (95% CI: 0.757-0.818), and 0.793 (95% CI: 0.758-0.831) for ccRCC, pRCC, and chRCC, respectively. Conclusions The developed deep learning model demonstrated robust performance in predicting the pathological subtypes of RCC, while the incorporated uncertainty emphasized the importance of understanding model confidence, which is crucial for assisting clinical decision-making for patients with renal tumors. Clinical relevance statement Our deep learning approach, integrated with uncertainty estimation, offers clinicians a dual advantage: accurate RCC subtype predictions complemented by diagnostic confidence references, promoting informed decision-making for patients with RCC.
Machine learning for the prediction of safe and biologically active organophosphorus molecules
Feb 21, 2023


Abstract:Drug discovery is a complex process with a large molecular space to be considered. By constraining the search space, the fragment-based drug design is an approach that can effectively sample the chemical space of interest. Here we propose a framework of Recurrent Neural Networks (RNN) with an attention model to sample the chemical space of organophosphorus molecules using the fragment-based approach. The framework is trained with a ZINC dataset that is screened for high druglikeness scores. The goal is to predict molecules with similar biological action modes as organophosphorus pesticides or chemical warfare agents yet less toxic to humans. The generated molecules contain a starting fragment of PO2F but have a bulky hydrocarbon side chain limiting its binding effectiveness to the targeted protein.
CHA2: CHemistry Aware Convex Hull Autoencoder Towards Inverse Molecular Design
Feb 21, 2023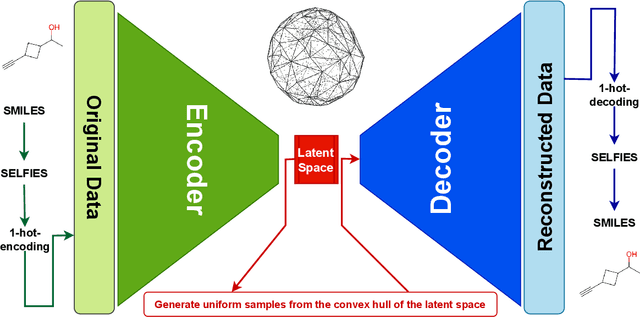
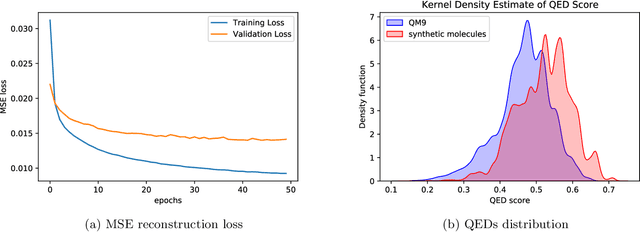


Abstract:Optimizing molecular design and discovering novel chemical structures to meet certain objectives, such as quantitative estimates of the drug-likeness score (QEDs), is NP-hard due to the vast combinatorial design space of discrete molecular structures, which makes it near impossible to explore the entire search space comprehensively to exploit de novo structures with properties of interest. To address this challenge, reducing the intractable search space into a lower-dimensional latent volume helps examine molecular candidates more feasibly via inverse design. Autoencoders are suitable deep learning techniques, equipped with an encoder that reduces the discrete molecular structure into a latent space and a decoder that inverts the search space back to the molecular design. The continuous property of the latent space, which characterizes the discrete chemical structures, provides a flexible representation for inverse design in order to discover novel molecules. However, exploring this latent space requires certain insights to generate new structures. We propose using a convex hall surrounding the top molecules in terms of high QEDs to ensnare a tight subspace in the latent representation as an efficient way to reveal novel molecules with high QEDs. We demonstrate the effectiveness of our suggested method by using the QM9 as a training dataset along with the Self- Referencing Embedded Strings (SELFIES) representation to calibrate the autoencoder in order to carry out the Inverse molecular design that leads to unfold novel chemical structure.
Deep Learning Approach for Dynamic Sampling for Multichannel Mass Spectrometry Imaging
Oct 24, 2022Abstract:Mass Spectrometry Imaging (MSI), using traditional rectilinear scanning, takes hours to days for high spatial resolution acquisitions. Given that most pixels within a sample's field of view are often neither relevant to underlying biological structures nor chemically informative, MSI presents as a prime candidate for integration with sparse and dynamic sampling algorithms. During a scan, stochastic models determine which locations probabilistically contain information critical to the generation of low-error reconstructions. Decreasing the number of required physical measurements thereby minimizes overall acquisition times. A Deep Learning Approach for Dynamic Sampling (DLADS), utilizing a Convolutional Neural Network (CNN) and encapsulating molecular mass intensity distributions within a third dimension, demonstrates a simulated 70% throughput improvement for Nanospray Desorption Electrospray Ionization (nano-DESI) MSI tissues. Evaluations are conducted between DLADS and a Supervised Learning Approach for Dynamic Sampling, with Least-Squares regression (SLADS-LS) and a Multi-Layer Perceptron (MLP) network (SLADS-Net). When compared with SLADS-LS, limited to a single m/z channel, as well as multichannel SLADS-LS and SLADS-Net, DLADS respectively improves regression performance by 36.7%, 7.0%, and 6.2%, resulting in gains to reconstruction quality of 6.0%, 2.1%, and 3.4% for acquisition of targeted m/z.
Training Overparametrized Neural Networks in Sublinear Time
Aug 09, 2022Abstract:The success of deep learning comes at a tremendous computational and energy cost, and the scalability of training massively overparametrized neural networks is becoming a real barrier to the progress of AI. Despite the popularity and low cost-per-iteration of traditional Backpropagation via gradient decent, SGD has prohibitive convergence rate in non-convex settings, both in theory and practice. To mitigate this cost, recent works have proposed to employ alternative (Newton-type) training methods with much faster convergence rate, albeit with higher cost-per-iteration. For a typical neural network with $m=\mathrm{poly}(n)$ parameters and input batch of $n$ datapoints in $\mathbb{R}^d$, the previous work of [Brand, Peng, Song, and Weinstein, ITCS'2021] requires $\sim mnd + n^3$ time per iteration. In this paper, we present a novel training method that requires only $m^{1-\alpha} n d + n^3$ amortized time in the same overparametrized regime, where $\alpha \in (0.01,1)$ is some fixed constant. This method relies on a new and alternative view of neural networks, as a set of binary search trees, where each iteration corresponds to modifying a small subset of the nodes in the tree. We believe this view would have further applications in the design and analysis of DNNs.
Sublinear Time Algorithm for Online Weighted Bipartite Matching
Aug 05, 2022Abstract:Online bipartite matching is a fundamental problem in online algorithms. The goal is to match two sets of vertices to maximize the sum of the edge weights, where for one set of vertices, each vertex and its corresponding edge weights appear in a sequence. Currently, in the practical recommendation system or search engine, the weights are decided by the inner product between the deep representation of a user and the deep representation of an item. The standard online matching needs to pay $nd$ time to linear scan all the $n$ items, computing weight (assuming each representation vector has length $d$), and then decide the matching based on the weights. However, in reality, the $n$ could be very large, e.g. in online e-commerce platforms. Thus, improving the time of computing weights is a problem of practical significance. In this work, we provide the theoretical foundation for computing the weights approximately. We show that, with our proposed randomized data structures, the weights can be computed in sublinear time while still preserving the competitive ratio of the matching algorithm.
Quasi-Direct Drive Actuation for a Lightweight Hip Exoskeleton with High Backdrivability and High Bandwidth
Apr 01, 2020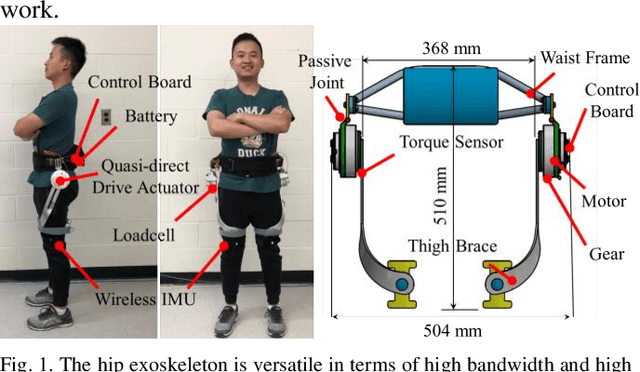
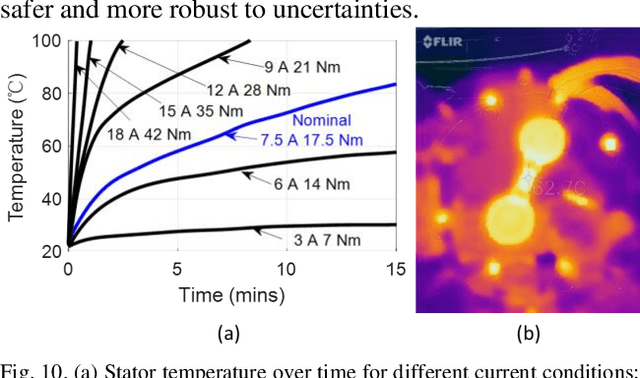
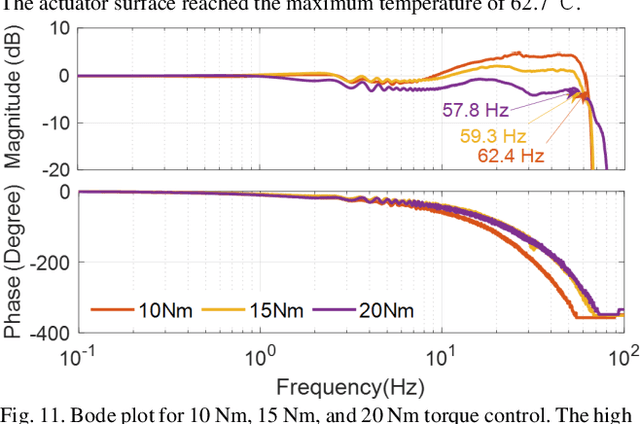
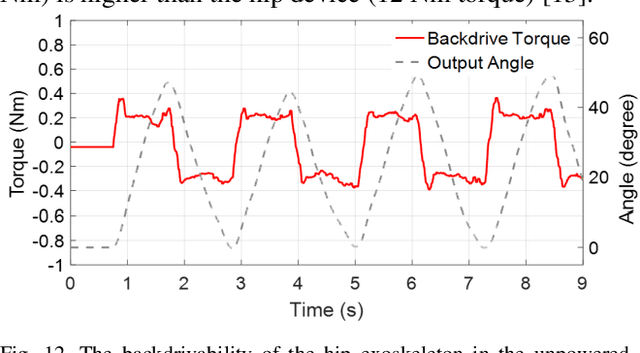
Abstract:High-performance actuators are crucial to enable mechanical versatility of lower-limb wearable robots, which are required to be lightweight, highly backdrivable, and with high bandwidth. State-of-the-art actuators, e.g., series elastic actuators (SEAs), have to compromise bandwidth to improve compliance (i.e., backdrivability). In this paper, we describe the design and human-robot interaction modeling of a portable hip exoskeleton based on our custom quasi-direct drive (QDD) actuation (i.e., a high torque density motor with low ratio gear). We also present a model-based performance benchmark comparison of representative actuators in terms of torque capability, control bandwidth, backdrivability, and force tracking accuracy. This paper aims to corroborate the underlying philosophy of "design for control", namely meticulous robot design can simplify control algorithms while ensuring high performance. Following this idea, we create a lightweight bilateral hip exoskeleton (overall mass is 3.4 kg) to reduce joint loadings during normal activities, including walking and squatting. Experimental results indicate that the exoskeleton is able to produce high nominal torque (17.5 Nm), high backdrivability (0.4 Nm backdrive torque), high bandwidth (62.4 Hz), and high control accuracy (1.09 Nm root mean square tracking error, i.e., 5.4% of the desired peak torque). Its controller is versatile to assist walking at different speeds (0.8-1.4 m/s) and squatting at 2 s cadence. This work demonstrates significant improvement in backdrivability and control bandwidth compared with state-of-the-art exoskeletons powered by the conventional actuation or SEA.
Spine-Inspired Continuum Soft Exoskeleton for Stoop Lifting Assistance
Jul 04, 2019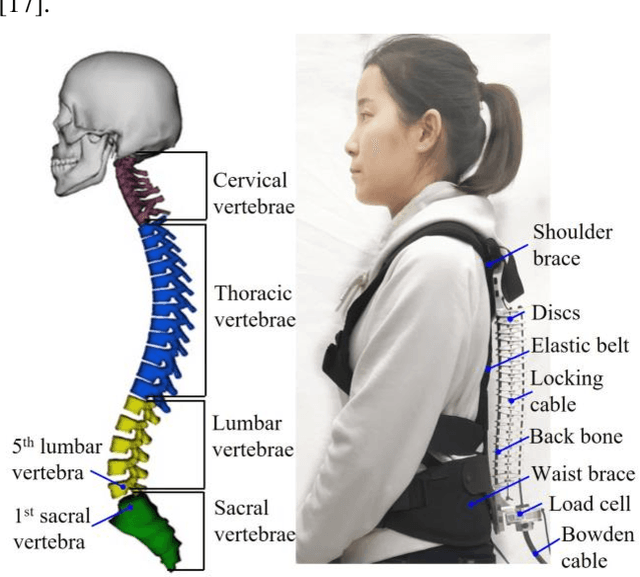
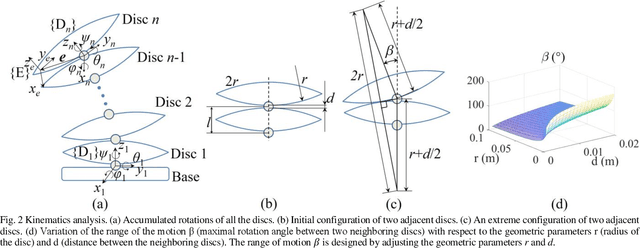
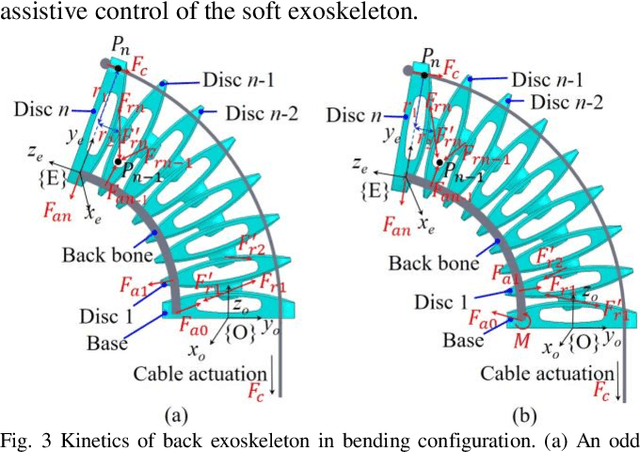
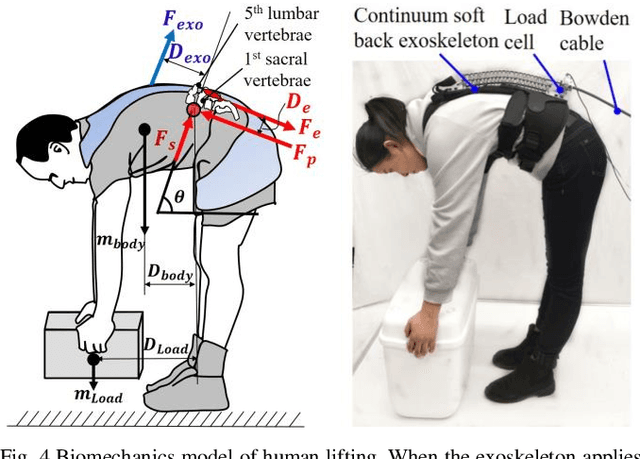
Abstract:Back injuries are the most prevalent work-related musculoskeletal disorders and represent a major cause of disability. Although innovations in wearable robots aim to alleviate this hazard, the majority of existing exoskeletons are obtrusive because the rigid linkage design limits natural movement, thus causing ergonomic risk. Moreover, these existing systems are typically only suitable for one type of movement assistance, not ubiquitous for a wide variety of activities. To fill in this gap, this paper presents a new wearable robot design approach continuum soft exoskeleton. This spine-inspired wearable robot is unobtrusive and assists both squat and stoops while not impeding walking motion. To tackle the challenge of the unique anatomy of spine that is inappropriate to be simplified as a single degree of freedom joint, our robot is conformal to human anatomy and it can reduce multiple types of forces along the human spine such as the spinae muscle force, shear, and compression force of the lumbar vertebrae. We derived kinematics and kinetics models of this mechanism and established an analytical biomechanics model of human-robot interaction. Quantitative analysis of disc compression force, disc shear force and muscle force was performed in simulation. We further developed a virtual impedance control strategy to deliver force control and compensate hysteresis of Bowden cable transmission. The feasibility of the prototype was experimentally tested on three healthy subjects. The root mean square error of force tracking is 6.63 N (3.3 % of the 200N peak force) and it demonstrated that it can actively control the stiffness to the desired value. This continuum soft exoskeleton represents a feasible solution with the potential to reduce back pain for multiple activities and multiple forces along the human spine.
* 8 pages, 13 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge