Jiaofen Nan
UPMAD-Net: A Brain Tumor Segmentation Network with Uncertainty Guidance and Adaptive Multimodal Feature Fusion
May 06, 2025Abstract:Background: Brain tumor segmentation has a significant impact on the diagnosis and treatment of brain tumors. Accurate brain tumor segmentation remains challenging due to their irregular shapes, vague boundaries, and high variability. Objective: We propose a brain tumor segmentation method that combines deep learning with prior knowledge derived from a region-growing algorithm. Methods: The proposed method utilizes a multi-scale feature fusion (MSFF) module and adaptive attention mechanisms (AAM) to extract multi-scale features and capture global contextual information. To enhance the model's robustness in low-confidence regions, the Monte Carlo Dropout (MC Dropout) strategy is employed for uncertainty estimation. Results: Extensive experiments demonstrate that the proposed method achieves superior performance on Brain Tumor Segmentation (BraTS) datasets, significantly outperforming various state-of-the-art methods. On the BraTS2021 dataset, the test Dice scores are 89.18% for Enhancing Tumor (ET) segmentation, 93.67% for Whole Tumor (WT) segmentation, and 91.23% for Tumor Core (TC) segmentation. On the BraTS2019 validation set, the validation Dice scores are 87.43%, 90.92%, and 90.40% for ET, WT, and TC segmentation, respectively. Ablation studies further confirmed the contribution of each module to segmentation accuracy, indicating that each component played a vital role in overall performance improvement. Conclusion: This study proposed a novel 3D brain tumor segmentation network based on the U-Net architecture. By incorporating the prior knowledge and employing the uncertainty estimation method, the robustness and performance were improved. The code for the proposed method is available at https://github.com/chenzhao2023/UPMAD_Net_BrainSeg.
Myocardial Region-guided Feature Aggregation Net for Automatic Coronary artery Segmentation and Stenosis Assessment using Coronary Computed Tomography Angiography
Apr 27, 2025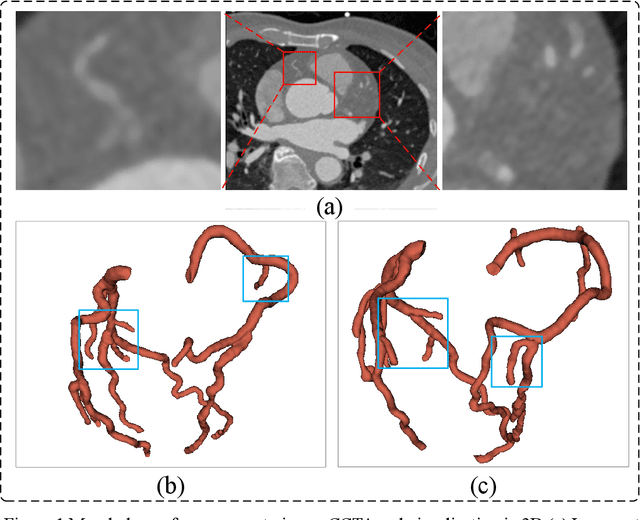
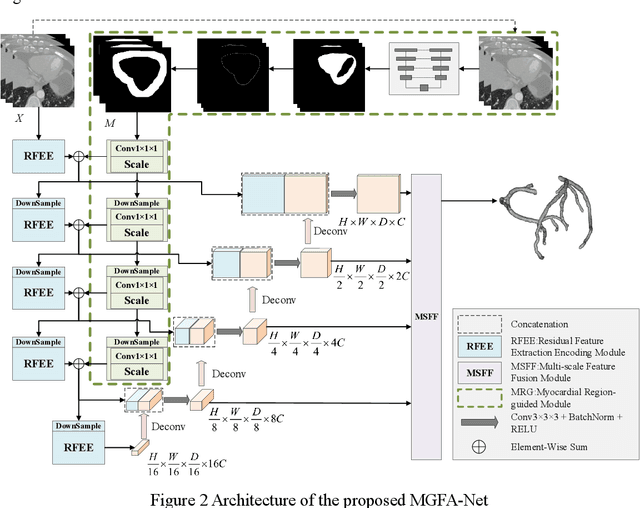
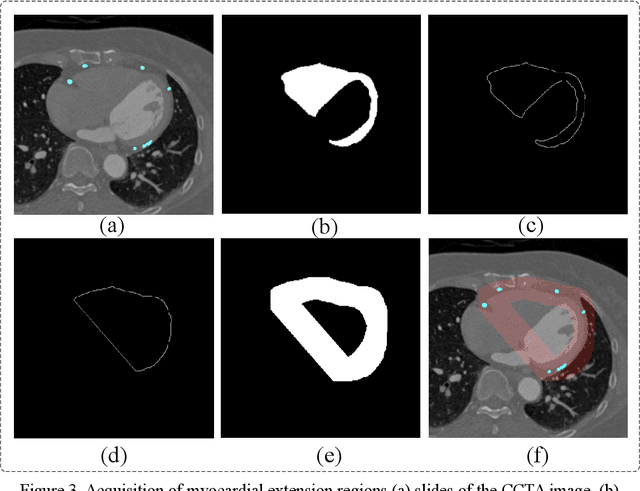
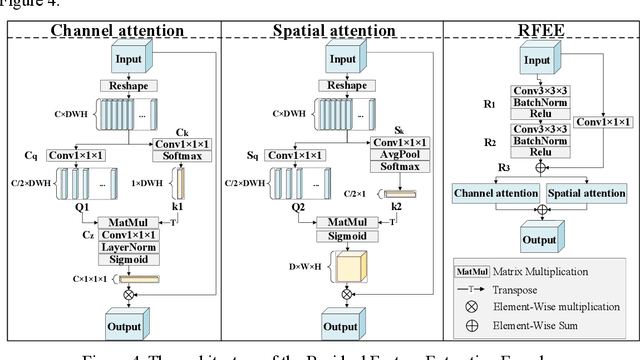
Abstract:Coronary artery disease (CAD) remains a leading cause of mortality worldwide, requiring accurate segmentation and stenosis detection using Coronary Computed Tomography angiography (CCTA). Existing methods struggle with challenges such as low contrast, morphological variability and small vessel segmentation. To address these limitations, we propose the Myocardial Region-guided Feature Aggregation Net, a novel U-shaped dual-encoder architecture that integrates anatomical prior knowledge to enhance robustness in coronary artery segmentation. Our framework incorporates three key innovations: (1) a Myocardial Region-guided Module that directs attention to coronary regions via myocardial contour expansion and multi-scale feature fusion, (2) a Residual Feature Extraction Encoding Module that combines parallel spatial channel attention with residual blocks to enhance local-global feature discrimination, and (3) a Multi-scale Feature Fusion Module for adaptive aggregation of hierarchical vascular features. Additionally, Monte Carlo dropout f quantifies prediction uncertainty, supporting clinical interpretability. For stenosis detection, a morphology-based centerline extraction algorithm separates the vascular tree into anatomical branches, enabling cross-sectional area quantification and stenosis grading. The superiority of MGFA-Net was demonstrated by achieving an Dice score of 85.04%, an accuracy of 84.24%, an HD95 of 6.1294 mm, and an improvement of 5.46% in true positive rate for stenosis detection compared to3D U-Net. The integrated segmentation-to-stenosis pipeline provides automated, clinically interpretable CAD assessment, bridging deep learning with anatomical prior knowledge for precision medicine. Our code is publicly available at http://github.com/chenzhao2023/MGFA_CCTA
MsMorph: An Unsupervised pyramid learning network for brain image registration
Oct 23, 2024Abstract:In the field of medical image analysis, image registration is a crucial technique. Despite the numerous registration models that have been proposed, existing methods still fall short in terms of accuracy and interpretability. In this paper, we present MsMorph, a deep learning-based image registration framework aimed at mimicking the manual process of registering image pairs to achieve more similar deformations, where the registered image pairs exhibit consistency or similarity in features. By extracting the feature differences between image pairs across various as-pects using gradients, the framework decodes semantic information at different scales and continuously compen-sates for the predicted deformation field, driving the optimization of parameters to significantly improve registration accuracy. The proposed method simulates the manual approach to registration, focusing on different regions of the image pairs and their neighborhoods to predict the deformation field between the two images, which provides strong interpretability. We compared several existing registration methods on two public brain MRI datasets, including LPBA and Mindboggle. The experimental results show that our method consistently outperforms state of the art in terms of metrics such as Dice score, Hausdorff distance, average symmetric surface distance, and non-Jacobian. The source code is publicly available at https://github.com/GaodengFan/MsMorph
Vision Transformer-based Multimodal Feature Fusion Network for Lymphoma Segmentation on PET/CT Images
Feb 04, 2024Abstract:Background: Diffuse large B-cell lymphoma (DLBCL) segmentation is a challenge in medical image analysis. Traditional segmentation methods for lymphoma struggle with the complex patterns and the presence of DLBCL lesions. Objective: We aim to develop an accurate method for lymphoma segmentation with 18F-Fluorodeoxyglucose positron emission tomography (PET) and computed tomography (CT) images. Methods: Our lymphoma segmentation approach combines a vision transformer with dual encoders, adeptly fusing PET and CT data via multimodal cross-attention fusion (MMCAF) module. In this study, PET and CT data from 165 DLBCL patients were analyzed. A 5-fold cross-validation was employed to evaluate the performance and generalization ability of our method. Ground truths were annotated by experienced nuclear medicine experts. We calculated the total metabolic tumor volume (TMTV) and performed a statistical analysis on our results. Results: The proposed method exhibited accurate performance in DLBCL lesion segmentation, achieving a Dice similarity coefficient of 0.9173$\pm$0.0071, a Hausdorff distance of 2.71$\pm$0.25mm, a sensitivity of 0.9462$\pm$0.0223, and a specificity of 0.9986$\pm$0.0008. Additionally, a Pearson correlation coefficient of 0.9030$\pm$0.0179 and an R-square of 0.8586$\pm$0.0173 were observed in TMTV when measured on manual annotation compared to our segmentation results. Conclusion: This study highlights the advantages of MMCAF and vision transformer for lymphoma segmentation using PET and CT, offering great promise for computer-aided lymphoma diagnosis and treatment.
A Robust Deep Learning Method with Uncertainty Estimation for the Pathological Classification of Renal Cell Carcinoma based on CT Images
Nov 12, 2023



Abstract:Objectives To develop and validate a deep learning-based diagnostic model incorporating uncertainty estimation so as to facilitate radiologists in the preoperative differentiation of the pathological subtypes of renal cell carcinoma (RCC) based on CT images. Methods Data from 668 consecutive patients, pathologically proven RCC, were retrospectively collected from Center 1. By using five-fold cross-validation, a deep learning model incorporating uncertainty estimation was developed to classify RCC subtypes into clear cell RCC (ccRCC), papillary RCC (pRCC), and chromophobe RCC (chRCC). An external validation set of 78 patients from Center 2 further evaluated the model's performance. Results In the five-fold cross-validation, the model's area under the receiver operating characteristic curve (AUC) for the classification of ccRCC, pRCC, and chRCC was 0.868 (95% CI: 0.826-0.923), 0.846 (95% CI: 0.812-0.886), and 0.839 (95% CI: 0.802-0.88), respectively. In the external validation set, the AUCs were 0.856 (95% CI: 0.838-0.882), 0.787 (95% CI: 0.757-0.818), and 0.793 (95% CI: 0.758-0.831) for ccRCC, pRCC, and chRCC, respectively. Conclusions The developed deep learning model demonstrated robust performance in predicting the pathological subtypes of RCC, while the incorporated uncertainty emphasized the importance of understanding model confidence, which is crucial for assisting clinical decision-making for patients with renal tumors. Clinical relevance statement Our deep learning approach, integrated with uncertainty estimation, offers clinicians a dual advantage: accurate RCC subtype predictions complemented by diagnostic confidence references, promoting informed decision-making for patients with RCC.
MLA-BIN: Model-level Attention and Batch-instance Style Normalization for Domain Generalization of Federated Learning on Medical Image Segmentation
Jun 29, 2023



Abstract:The privacy protection mechanism of federated learning (FL) offers an effective solution for cross-center medical collaboration and data sharing. In multi-site medical image segmentation, each medical site serves as a client of FL, and its data naturally forms a domain. FL supplies the possibility to improve the performance of seen domains model. However, there is a problem of domain generalization (DG) in the actual de-ployment, that is, the performance of the model trained by FL in unseen domains will decrease. Hence, MLA-BIN is proposed to solve the DG of FL in this study. Specifically, the model-level attention module (MLA) and batch-instance style normalization (BIN) block were designed. The MLA represents the unseen domain as a linear combination of seen domain models. The atten-tion mechanism is introduced for the weighting coefficient to obtain the optimal coefficient ac-cording to the similarity of inter-domain data features. MLA enables the global model to gen-eralize to unseen domain. In the BIN block, batch normalization (BN) and instance normalization (IN) are combined to perform the shallow layers of the segmentation network for style normali-zation, solving the influence of inter-domain image style differences on DG. The extensive experimental results of two medical image seg-mentation tasks demonstrate that the proposed MLA-BIN outperforms state-of-the-art methods.
A new method incorporating deep learning with shape priors for left ventricular segmentation in myocardial perfusion SPECT images
Jun 07, 2022



Abstract:Background: The assessment of left ventricular (LV) function by myocardial perfusion SPECT (MPS) relies on accurate myocardial segmentation. The purpose of this paper is to develop and validate a new method incorporating deep learning with shape priors to accurately extract the LV myocardium for automatic measurement of LV functional parameters. Methods: A segmentation architecture that integrates a three-dimensional (3D) V-Net with a shape deformation module was developed. Using the shape priors generated by a dynamic programming (DP) algorithm, the model output was then constrained and guided during the model training for quick convergence and improved performance. A stratified 5-fold cross-validation was used to train and validate our models. Results: Results of our proposed method agree well with those from the ground truth. Our proposed model achieved a Dice similarity coefficient (DSC) of 0.9573(0.0244), 0.9821(0.0137), and 0.9903(0.0041), a Hausdorff distances (HD) of 6.7529(2.7334) mm, 7.2507(3.1952) mm, and 7.6121(3.0134) mm in extracting the endocardium, myocardium, and epicardium, respectively. Conclusion: Our proposed method achieved a high accuracy in extracting LV myocardial contours and assessing LV function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge