Zhixin Jiang
FedDA-TSformer: Federated Domain Adaptation with Vision TimeSformer for Left Ventricle Segmentation on Gated Myocardial Perfusion SPECT Image
Feb 23, 2025Abstract:Background and Purpose: Functional assessment of the left ventricle using gated myocardial perfusion (MPS) single-photon emission computed tomography relies on the precise extraction of the left ventricular contours while simultaneously ensuring the security of patient data. Methods: In this paper, we introduce the integration of Federated Domain Adaptation with TimeSformer, named 'FedDA-TSformer' for left ventricle segmentation using MPS. FedDA-TSformer captures spatial and temporal features in gated MPS images, leveraging spatial attention, temporal attention, and federated learning for improved domain adaptation while ensuring patient data security. In detail, we employed Divide-Space-Time-Attention mechanism to extract spatio-temporal correlations from the multi-centered MPS datasets, ensuring that predictions are spatio-temporally consistent. To achieve domain adaptation, we align the model output on MPS from three different centers using local maximum mean discrepancy (LMMD) loss. This approach effectively addresses the dual requirements of federated learning and domain adaptation, enhancing the model's performance during training with multi-site datasets while ensuring the protection of data from different hospitals. Results: Our FedDA-TSformer was trained and evaluated using MPS datasets collected from three hospitals, comprising a total of 150 subjects. Each subject's cardiac cycle was divided into eight gates. The model achieved Dice Similarity Coefficients (DSC) of 0.842 and 0.907 for left ventricular (LV) endocardium and epicardium segmentation, respectively. Conclusion: Our proposed FedDA-TSformer model addresses the challenge of multi-center generalization, ensures patient data privacy protection, and demonstrates effectiveness in left ventricular (LV) segmentation.
Automatic reorientation by deep learning to generate short axis SPECT myocardial perfusion images
Aug 07, 2022
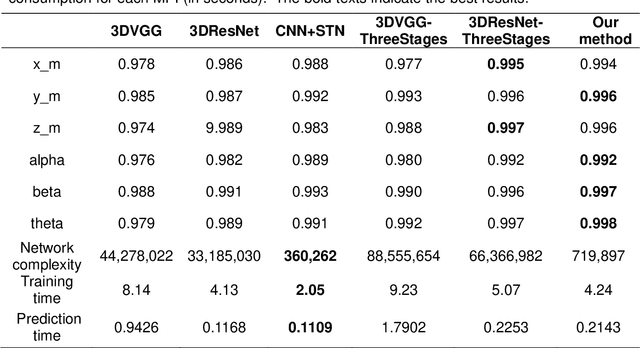
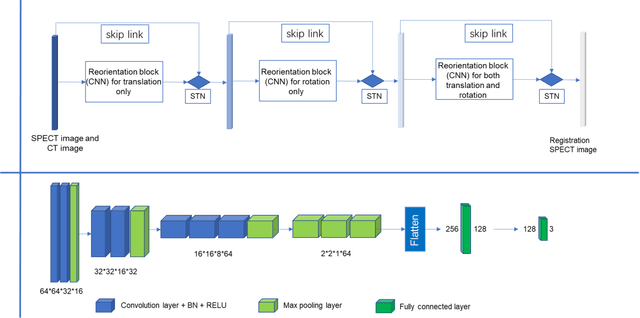

Abstract:Single photon emission computed tomography (SPECT) myocardial perfusion images (MPI) can be displayed both in traditional short-axis (SA) cardiac planes and polar maps for interpretation and quantification. It is essential to reorient the reconstructed transaxial SPECT MPI into standard SA slices. This study is aimed to develop a deep-learning-based approach for automatic reorientation of MPI. Methods: A total of 254 patients were enrolled, including 228 stress SPECT MPIs and 248 rest SPECT MPIs. Five-fold cross-validation with 180 stress and 201 rest MPIs was used for training and internal validation; the remaining images were used for testing. The rigid transformation parameters (translation and rotation) from manual reorientation were annotated by an experienced operator and used as the ground truth. A convolutional neural network (CNN) was designed to predict the transformation parameters. Then, the derived transform was applied to the grid generator and sampler in spatial transformer network (STN) to generate the reoriented image. A loss function containing mean absolute errors for translation and mean square errors for rotation was employed. A three-stage optimization strategy was adopted for model optimization: 1) optimize the translation parameters while fixing the rotation parameters; 2) optimize rotation parameters while fixing the translation parameters; 3) optimize both translation and rotation parameters together.
A new method incorporating deep learning with shape priors for left ventricular segmentation in myocardial perfusion SPECT images
Jun 07, 2022



Abstract:Background: The assessment of left ventricular (LV) function by myocardial perfusion SPECT (MPS) relies on accurate myocardial segmentation. The purpose of this paper is to develop and validate a new method incorporating deep learning with shape priors to accurately extract the LV myocardium for automatic measurement of LV functional parameters. Methods: A segmentation architecture that integrates a three-dimensional (3D) V-Net with a shape deformation module was developed. Using the shape priors generated by a dynamic programming (DP) algorithm, the model output was then constrained and guided during the model training for quick convergence and improved performance. A stratified 5-fold cross-validation was used to train and validate our models. Results: Results of our proposed method agree well with those from the ground truth. Our proposed model achieved a Dice similarity coefficient (DSC) of 0.9573(0.0244), 0.9821(0.0137), and 0.9903(0.0041), a Hausdorff distances (HD) of 6.7529(2.7334) mm, 7.2507(3.1952) mm, and 7.6121(3.0134) mm in extracting the endocardium, myocardium, and epicardium, respectively. Conclusion: Our proposed method achieved a high accuracy in extracting LV myocardial contours and assessing LV function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge