Han Peng
ForesightKV: Optimizing KV Cache Eviction for Reasoning Models by Learning Long-Term Contribution
Feb 03, 2026Abstract:Recently, large language models (LLMs) have shown remarkable reasoning abilities by producing long reasoning traces. However, as the sequence length grows, the key-value (KV) cache expands linearly, incurring significant memory and computation costs. Existing KV cache eviction methods mitigate this issue by discarding less important KV pairs, but often fail to capture complex KV dependencies, resulting in performance degradation. To better balance efficiency and performance, we introduce ForesightKV, a training-based KV cache eviction framework that learns to predict which KV pairs to evict during long-text generations. We first design the Golden Eviction algorithm, which identifies the optimal eviction KV pairs at each step using future attention scores. These traces and the scores at each step are then distilled via supervised training with a Pairwise Ranking Loss. Furthermore, we formulate cache eviction as a Markov Decision Process and apply the GRPO algorithm to mitigate the significant language modeling loss increase on low-entropy tokens. Experiments on AIME2024 and AIME2025 benchmarks of three reasoning models demonstrate that ForesightKV consistently outperforms prior methods under only half the cache budget, while benefiting synergistically from both supervised and reinforcement learning approaches.
CAFE: Retrieval Head-based Coarse-to-Fine Information Seeking to Enhance Multi-Document QA Capability
May 15, 2025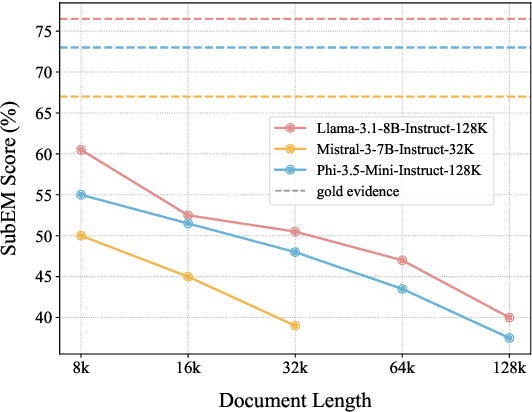


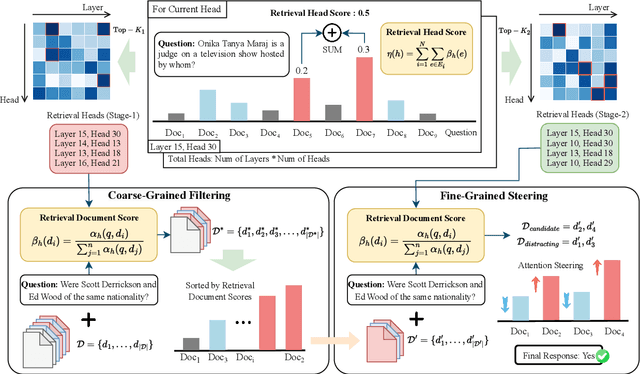
Abstract:Advancements in Large Language Models (LLMs) have extended their input context length, yet they still struggle with retrieval and reasoning in long-context inputs. Existing methods propose to utilize the prompt strategy and retrieval head to alleviate this limitation. However, they still face challenges in balancing retrieval precision and recall, impacting their efficacy in answering questions. To address this, we introduce $\textbf{CAFE}$, a two-stage coarse-to-fine method to enhance multi-document question-answering capacities. By gradually eliminating the negative impacts of background and distracting documents, CAFE makes the responses more reliant on the evidence documents. Initially, a coarse-grained filtering method leverages retrieval heads to identify and rank relevant documents. Then, a fine-grained steering method guides attention to the most relevant content. Experiments across benchmarks show CAFE outperforms baselines, achieving up to 22.1% and 13.7% SubEM improvement over SFT and RAG methods on the Mistral model, respectively.
Domain-Specific Pruning of Large Mixture-of-Experts Models with Few-shot Demonstrations
Apr 09, 2025



Abstract:Mixture-of-Experts (MoE) models achieve a favorable trade-off between performance and inference efficiency by activating only a subset of experts. However, the memory overhead of storing all experts remains a major limitation, especially in large-scale MoE models such as DeepSeek-R1 (671B). In this study, we investigate domain specialization and expert redundancy in large-scale MoE models and uncover a consistent behavior we term few-shot expert localization, with only a few demonstrations, the model consistently activates a sparse and stable subset of experts. Building on this observation, we propose a simple yet effective pruning framework, EASY-EP, that leverages a few domain-specific demonstrations to identify and retain only the most relevant experts. EASY-EP comprises two key components: output-aware expert importance assessment and expert-level token contribution estimation. The former evaluates the importance of each expert for the current token by considering the gating scores and magnitudes of the outputs of activated experts, while the latter assesses the contribution of tokens based on representation similarities after and before routed experts. Experiments show that our method can achieve comparable performances and $2.99\times$ throughput under the same memory budget with full DeepSeek-R1 with only half the experts. Our code is available at https://github.com/RUCAIBox/EASYEP.
M$^{3}$-20M: A Large-Scale Multi-Modal Molecule Dataset for AI-driven Drug Design and Discovery
Dec 08, 2024



Abstract:This paper introduces M$^{3}$-20M, a large-scale Multi-Modal Molecular dataset that contains over 20 million molecules. Designed to support AI-driven drug design and discovery, M$^{3}$-20M is 71 times more in the number of molecules than the largest existing dataset, providing an unprecedented scale that can highly benefit training or fine-tuning large (language) models with superior performance for drug design and discovery. This dataset integrates one-dimensional SMILES, two-dimensional molecular graphs, three-dimensional molecular structures, physicochemical properties, and textual descriptions collected through web crawling and generated by using GPT-3.5, offering a comprehensive view of each molecule. To demonstrate the power of M$^{3}$-20M in drug design and discovery, we conduct extensive experiments on two key tasks: molecule generation and molecular property prediction, using large language models including GLM4, GPT-3.5, and GPT-4. Our experimental results show that M$^{3}$-20M can significantly boost model performance in both tasks. Specifically, it enables the models to generate more diverse and valid molecular structures and achieve higher property prediction accuracy than the existing single-modal datasets, which validates the value and potential of M$^{3}$-20M in supporting AI-driven drug design and discovery. The dataset is available at \url{https://github.com/bz99bz/M-3}.
LLMBox: A Comprehensive Library for Large Language Models
Jul 08, 2024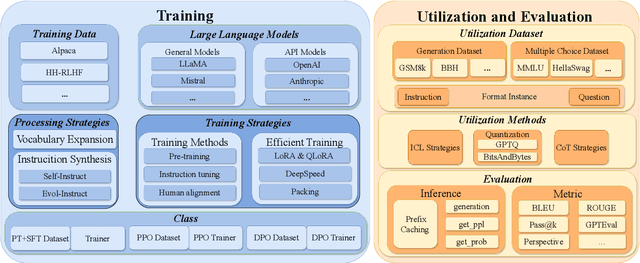
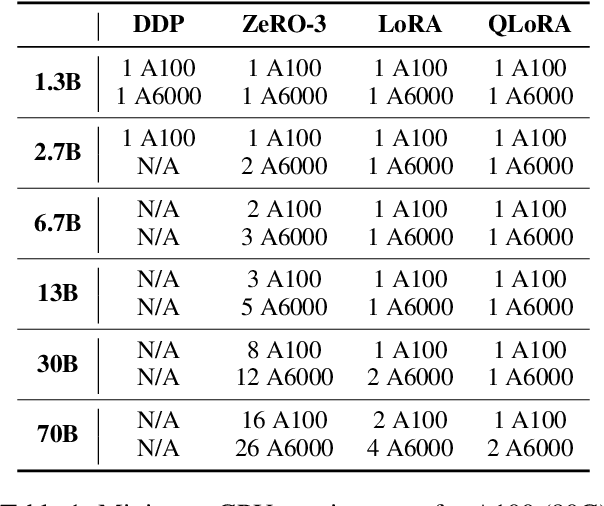
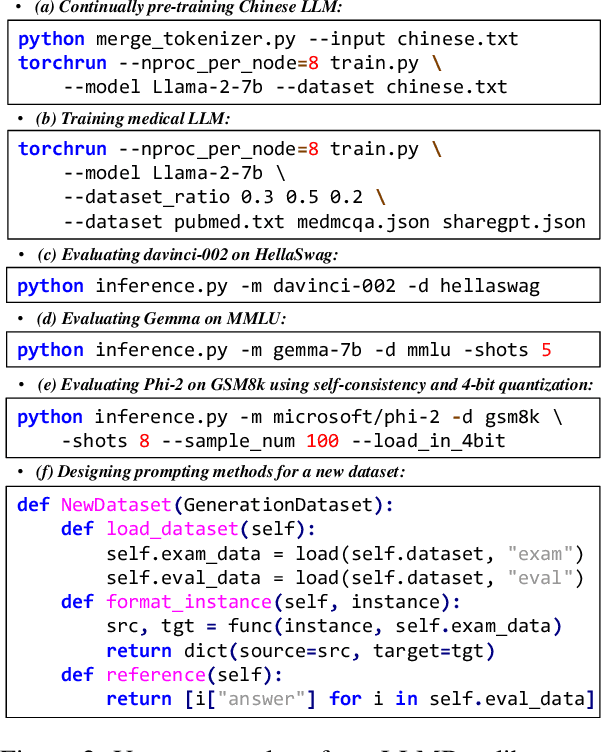
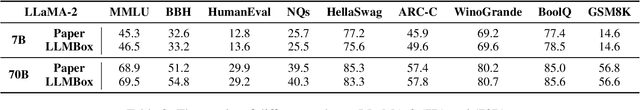
Abstract:To facilitate the research on large language models (LLMs), this paper presents a comprehensive and unified library, LLMBox, to ease the development, use, and evaluation of LLMs. This library is featured with three main merits: (1) a unified data interface that supports the flexible implementation of various training strategies, (2) a comprehensive evaluation that covers extensive tasks, datasets, and models, and (3) more practical consideration, especially on user-friendliness and efficiency. With our library, users can easily reproduce existing methods, train new models, and conduct comprehensive performance comparisons. To rigorously test LLMBox, we conduct extensive experiments in a diverse coverage of evaluation settings, and experimental results demonstrate the effectiveness and efficiency of our library in supporting various implementations related to LLMs. The detailed introduction and usage guidance can be found at https://github.com/RUCAIBox/LLMBox.
A Unified Framework for Joint Mobility Prediction and Object Profiling of Drones in UAV Networks
Jul 31, 2018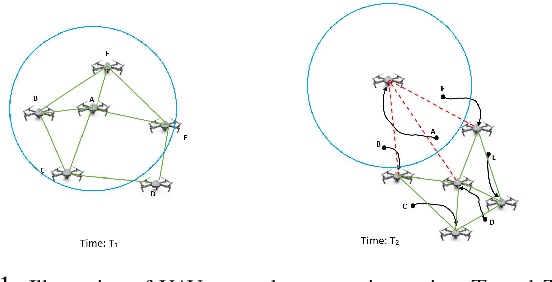
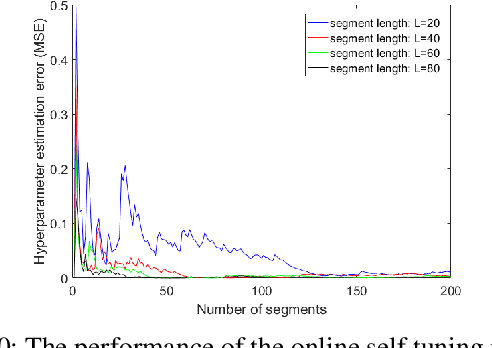
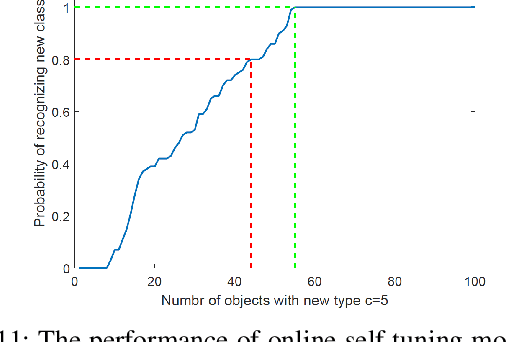
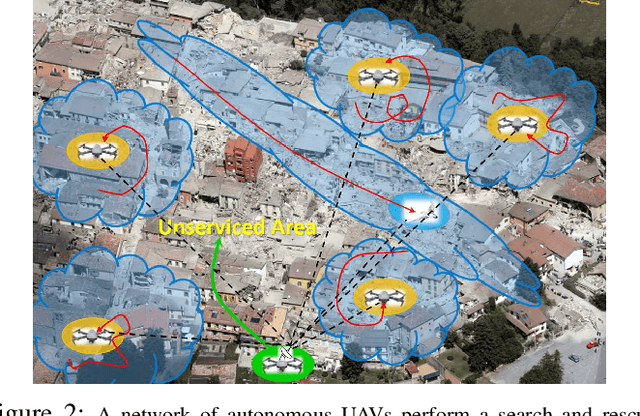
Abstract:In recent years, using a network of autonomous and cooperative unmanned aerial vehicles (UAVs) without command and communication from the ground station has become more imperative, in particular in search-and-rescue operations, disaster management, and other applications where human intervention is limited. In such scenarios, UAVs can make more efficient decisions if they acquire more information about the mobility, sensing and actuation capabilities of their neighbor nodes. In this paper, we develop an unsupervised online learning algorithm for joint mobility prediction and object profiling of UAVs to facilitate control and communication protocols. The proposed method not only predicts the future locations of the surrounding flying objects, but also classifies them into different groups with similar levels of maneuverability (e.g. rotatory, and fixed-wing UAVs) without prior knowledge about these classes. This method is flexible in admitting new object types with unknown mobility profiles, thereby applicable to emerging flying Ad-hoc networks with heterogeneous nodes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge