Guoan Zheng
Department of Biomedical Engineering, University of Connecticut, Storrs, CT, USA
Deep-ultraviolet ptychographic pocket-scope (DART): mesoscale lensless molecular imaging with label-free spectroscopic contrast
Nov 08, 2025Abstract:The mesoscale characterization of biological specimens has traditionally required compromises between resolution, field-of-view, depth-of-field, and molecular specificity, with most approaches relying on external labels. Here we present the Deep-ultrAviolet ptychogRaphic pockeT-scope (DART), a handheld platform that transforms label-free molecular imaging through intrinsic deep-ultraviolet spectroscopic contrast. By leveraging biomolecules' natural absorption fingerprints and combining them with lensless ptychographic microscopy, DART resolves down to 308-nm linewidths across centimeter-scale areas while maintaining millimeter-scale depth-of-field. The system's virtual error-bin methodology effectively eliminates artifacts from limited temporal coherence and other optical imperfections, enabling high-fidelity molecular imaging without lenses. Through differential spectroscopic imaging at deep-ultraviolet wavelengths, DART quantitatively maps nucleic acid and protein distributions with femtogram sensitivity, providing an intrinsic basis for explainable virtual staining. We demonstrate DART's capabilities through molecular imaging of tissue sections, cytopathology specimens, blood cells, and neural populations, revealing detailed molecular contrast without external labels. The combination of high-resolution molecular mapping and broad mesoscale imaging in a portable platform opens new possibilities from rapid clinical diagnostics, tissue analysis, to biological characterization in space exploration.
Video-rate gigapixel ptychography via space-time neural field representations
Nov 08, 2025Abstract:Achieving gigapixel space-bandwidth products (SBP) at video rates represents a fundamental challenge in imaging science. Here we demonstrate video-rate ptychography that overcomes this barrier by exploiting spatiotemporal correlations through neural field representations. Our approach factorizes the space-time volume into low-rank spatial and temporal features, transforming SBP scaling from sequential measurements to efficient correlation extraction. The architecture employs dual networks for decoding real and imaginary field components, avoiding phase-wrapping discontinuities plagued in amplitude-phase representations. A gradient-domain loss on spatial derivatives ensures robust convergence. We demonstrate video-rate gigapixel imaging with centimeter-scale coverage while resolving 308-nm linewidths. Validations span from monitoring sample dynamics of crystals, bacteria, stem cells, microneedle to characterizing time-varying probes in extreme ultraviolet experiments, demonstrating versatility across wavelengths. By transforming temporal variations from a constraint into exploitable correlations, we establish that gigapixel video is tractable with single-sensor measurements, making ptychography a high-throughput sensing tool for monitoring mesoscale dynamics without lenses.
Multiscale aperture synthesis imager
Nov 08, 2025Abstract:Synthetic aperture imaging has enabled breakthrough observations from radar to astronomy. However, optical implementation remains challenging due to stringent wavefield synchronization requirements among multiple receivers. Here we present the multiscale aperture synthesis imager (MASI), which utilizes parallelism to break complex optical challenges into tractable sub-problems. MASI employs a distributed array of coded sensors that operate independently yet coherently to surpass the diffraction limit of single receiver. It combines the propagated wavefields from individual sensors through a computational phase synchronization scheme, eliminating the need for overlapping measurement regions to establish phase coherence. Light diffraction in MASI naturally expands the imaging field, generating phase-contrast visualizations that are substantially larger than sensor dimensions. Without using lenses, MASI resolves sub-micron features at ultralong working distances and reconstructs 3D shapes over centimeter-scale fields. MASI transforms the intractable optical synchronization problem into a computational one, enabling practical deployment of scalable synthetic aperture systems at optical wavelengths.
Ptychographic non-line-of-sight imaging for depth-resolved visualization of hidden objects
May 17, 2024



Abstract:Non-line-of-sight (NLOS) imaging enables the visualization of objects hidden from direct view, with applications in surveillance, remote sensing, and light detection and ranging. Here, we introduce a NLOS imaging technique termed ptychographic NLOS (pNLOS), which leverages coded ptychography for depth-resolved imaging of obscured objects. Our approach involves scanning a laser spot on a wall to illuminate the hidden objects in an obscured region. The reflected wavefields from these objects then travel back to the wall, get modulated by the wall's complex-valued profile, and the resulting diffraction patterns are captured by a camera. By modulating the object wavefields, the wall surface serves the role of the coded layer as in coded ptychography. As we scan the laser spot to different positions, the reflected object wavefields on the wall translate accordingly, with the shifts varying for objects at different depths. This translational diversity enables the acquisition of a set of modulated diffraction patterns referred to as a ptychogram. By processing the ptychogram, we recover both the objects at different depths and the modulation profile of the wall surface. Experimental results demonstrate high-resolution, high-fidelity imaging of hidden objects, showcasing the potential of pNLOS for depth-aware vision beyond the direct line of sight.
Sparsity-regularized coded ptychography for robust and efficient lensless microscopy on a chip
Sep 24, 2023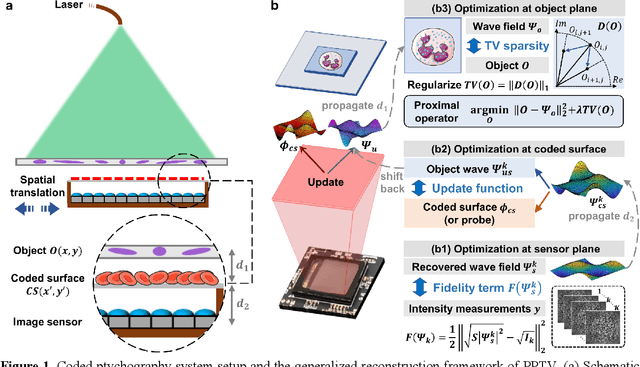

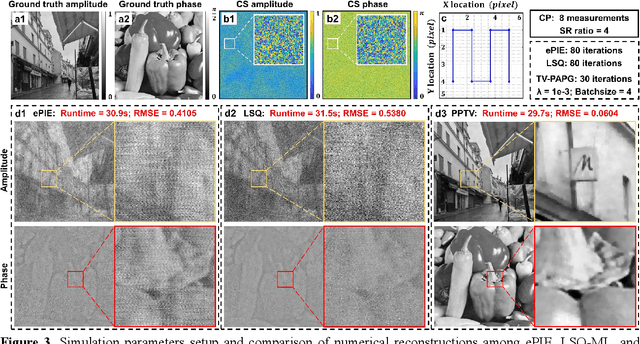

Abstract:In ptychographic imaging, the trade-off between the number of acquisitions and the resultant imaging quality presents a complex optimization problem. Increasing the number of acquisitions typically yields reconstructions with higher spatial resolution and finer details. Conversely, a reduction in measurement frequency often compromises the quality of the reconstructed images, manifesting as increased noise and coarser details. To address this challenge, we employ sparsity priors to reformulate the ptychographic reconstruction task as a total variation regularized optimization problem. We introduce a new computational framework, termed the ptychographic proximal total-variation (PPTV) solver, designed to integrate into existing ptychography settings without necessitating hardware modifications. Through comprehensive numerical simulations, we validate that PPTV-driven coded ptychography is capable of producing highly accurate reconstructions with a minimal set of eight intensity measurements. Convergence analysis further substantiates the robustness, stability, and computational feasibility of the proposed PPTV algorithm. Experimental results obtained from optical setups unequivocally demonstrate that the PPTV algorithm facilitates high-throughput, high-resolution imaging while significantly reducing the measurement burden. These findings indicate that the PPTV algorithm has the potential to substantially mitigate the resource-intensive requirements traditionally associated with high-quality ptychographic imaging, thereby offering a pathway toward the development of more compact and efficient ptychographic microscopy systems.
Digital staining in optical microscopy using deep learning -- a review
Mar 14, 2023Abstract:Until recently, conventional biochemical staining had the undisputed status as well-established benchmark for most biomedical problems related to clinical diagnostics, fundamental research and biotechnology. Despite this role as gold-standard, staining protocols face several challenges, such as a need for extensive, manual processing of samples, substantial time delays, altered tissue homeostasis, limited choice of contrast agents for a given sample, 2D imaging instead of 3D tomography and many more. Label-free optical technologies, on the other hand, do not rely on exogenous and artificial markers, by exploiting intrinsic optical contrast mechanisms, where the specificity is typically less obvious to the human observer. Over the past few years, digital staining has emerged as a promising concept to use modern deep learning for the translation from optical contrast to established biochemical contrast of actual stainings. In this review article, we provide an in-depth analysis of the current state-of-the-art in this field, suggest methods of good practice, identify pitfalls and challenges and postulate promising advances towards potential future implementations and applications.
Ptychographic sensor for large-scale lensless microbial monitoring with high spatiotemporal resolution
Dec 15, 2021
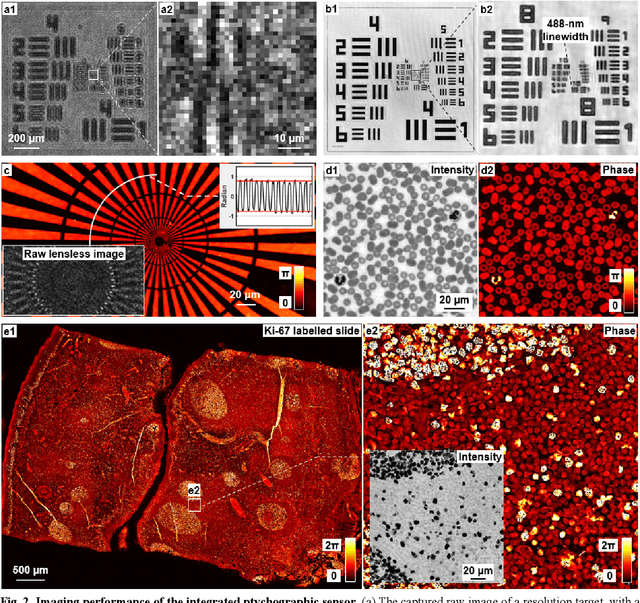

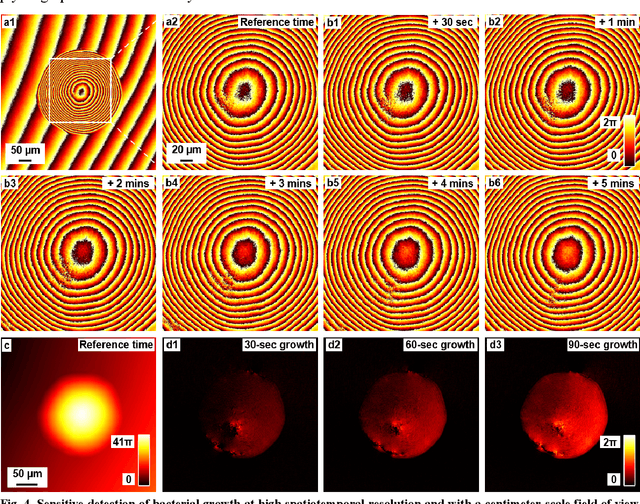
Abstract:Traditional microbial detection methods often rely on the overall property of microbial cultures and cannot resolve individual growth event at high spatiotemporal resolution. As a result, they require bacteria to grow to confluence and then interpret the results. Here, we demonstrate the application of an integrated ptychographic sensor for lensless cytometric analysis of microbial cultures over a large scale and with high spatiotemporal resolution. The reported device can be placed within a regular incubator or used as a standalone incubating unit for long-term microbial monitoring. For longitudinal study where massive data are acquired at sequential time points, we report a new temporal-similarity constraint to increase the temporal resolution of ptychographic reconstruction by 7-fold. With this strategy, the reported device achieves a centimeter-scale field of view, a half-pitch spatial resolution of 488 nm, and a temporal resolution of 15-second intervals. For the first time, we report the direct observation of bacterial growth in a 15-second interval by tracking the phase wraps of the recovered images, with high phase sensitivity like that in interferometric measurements. We also characterize cell growth via longitudinal dry mass measurement and perform rapid bacterial detection at low concentrations. For drug-screening application, we demonstrate proof-of-concept antibiotic susceptibility testing and perform single-cell analysis of antibiotic-induced filamentation. The combination of high phase sensitivity, high spatiotemporal resolution, and large field of view is unique among existing microscopy techniques. As a quantitative and miniaturized platform, it can improve studies with microorganisms and other biospecimens at resource-limited settings.
High-throughput lensless whole slide imaging via continuous height-varying modulation of tilted sensor
Sep 28, 2021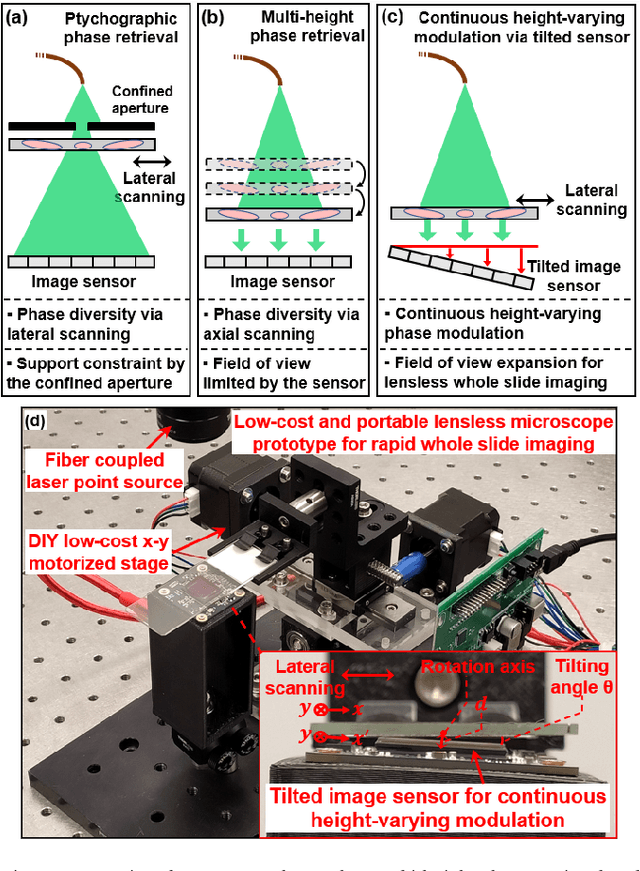
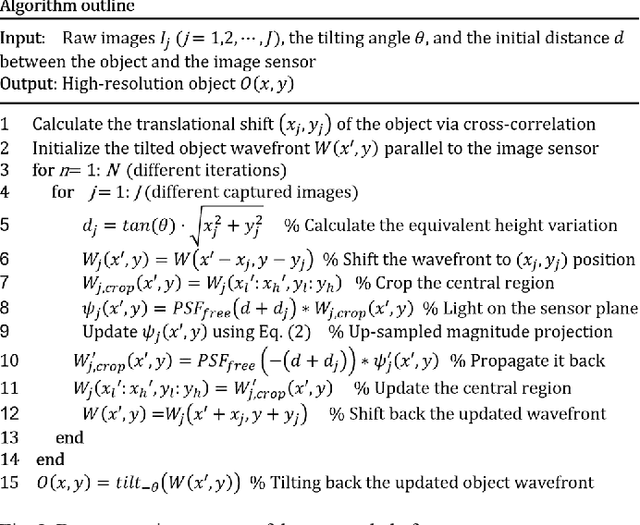

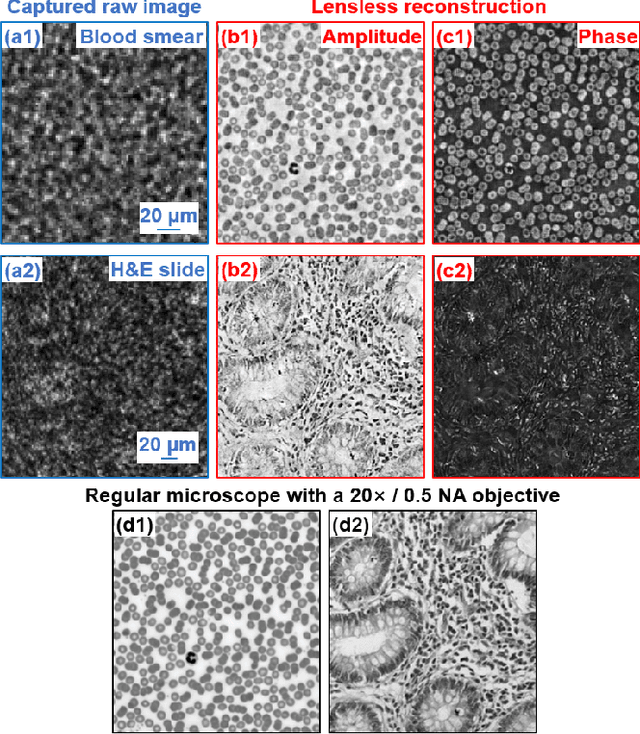
Abstract:We report a new lensless microscopy configuration by integrating the concepts of transverse translational ptychography and defocus multi-height phase retrieval. In this approach, we place a tilted image sensor under the specimen for linearly-increasing phase modulation along one lateral direction. Similar to the operation of ptychography, we laterally translate the specimen and acquire the diffraction images for reconstruction. Since the axial distance between the specimen and the sensor varies at different lateral positions, laterally translating the specimen effectively introduces defocus multi-height measurements while eliminating axial scanning. Lateral translation further introduces sub-pixel shift for pixel super-resolution imaging and naturally expands the field of view for rapid whole slide imaging. We show that the equivalent height variation can be precisely estimated from the lateral shift of the specimen, thereby addressing the challenge of precise axial positioning in conventional multi-height phase retrieval. Using a sensor with a 1.67-micron pixel size, our low-cost and field-portable prototype can resolve 690-nm linewidth on the resolution target. We show that a whole slide image of a blood smear with a 120-mm^2 field of view can be acquired in 18 seconds. We also demonstrate accurate automatic white blood cell counting from the recovered image. The reported approach may provide a turnkey solution for addressing point-of-care- and telemedicine-related challenges.
Plug-and-play optimization for pixel super-resolution phase retrieval
May 31, 2021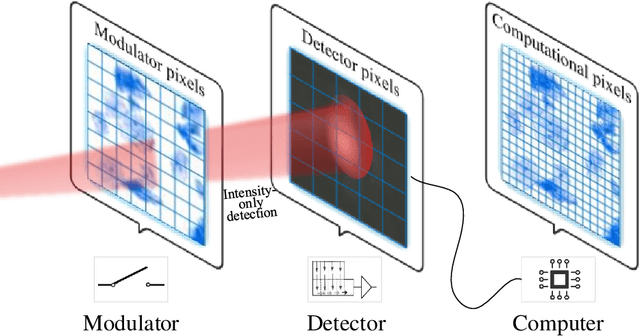

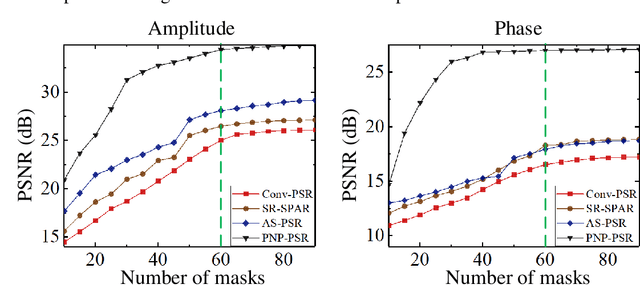

Abstract:In order to increase signal-to-noise ratio in measurement, most imaging detectors sacrifice resolution to increase pixel size in confined area. Although the pixel super-resolution technique (PSR) enables resolution enhancement in such as digital holographic imaging, it suffers from unsatisfied reconstruction quality. In this work, we report a high-fidelity plug-and-play optimization method for PSR phase retrieval, termed as PNP-PSR. It decomposes PSR reconstruction into independent sub-problems based on the generalized alternating projection framework. An alternating projection operator and an enhancing neural network are derived to tackle the measurement fidelity and statistical prior regularization, respectively. In this way, PNP-PSR incorporates the advantages of individual operators, achieving both high efficiency and noise robustness. We compare PNP-PSR with the existing PSR phase retrieval algorithms with a series of simulations and experiments, and PNP-PSR outperforms the existing algorithms with as much as 11dB on PSNR. The enhanced imaging fidelity enables one-order-of-magnitude higher cell counting precision.
Blind deblurring for microscopic pathology images using deep learning networks
Nov 24, 2020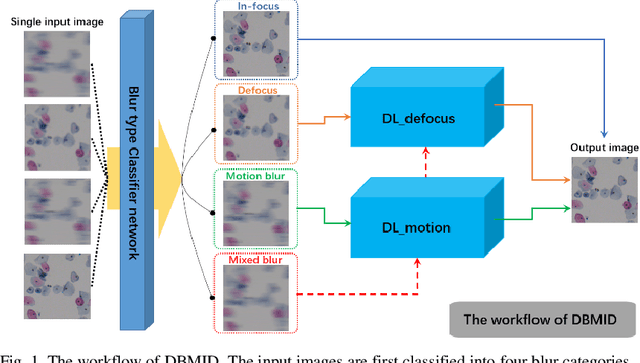
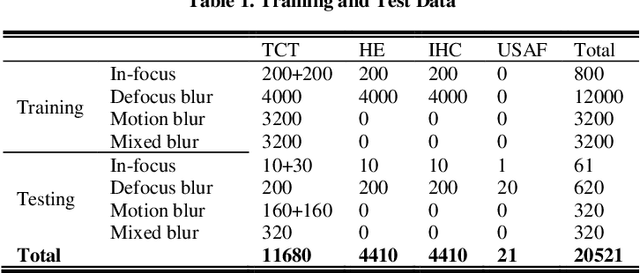
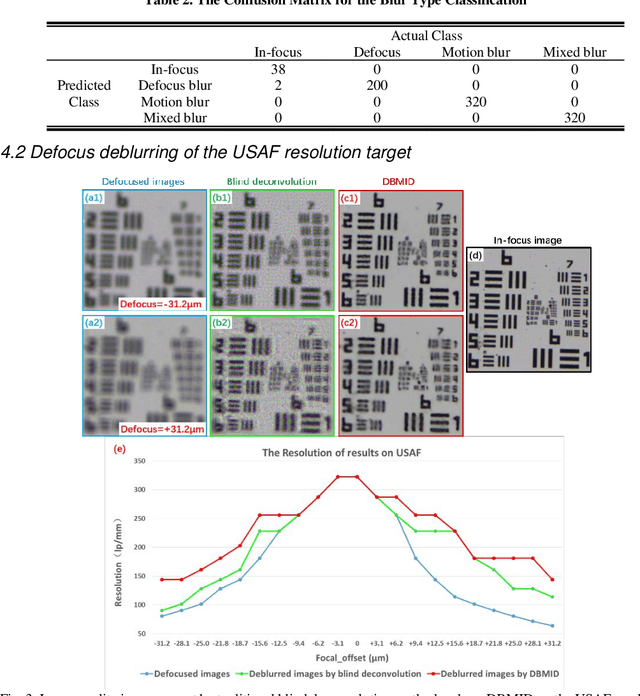
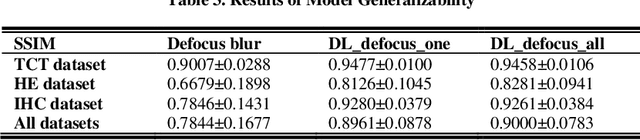
Abstract:Artificial Intelligence (AI)-powered pathology is a revolutionary step in the world of digital pathology and shows great promise to increase both diagnosis accuracy and efficiency. However, defocus and motion blur can obscure tissue or cell characteristics hence compromising AI algorithms'accuracy and robustness in analyzing the images. In this paper, we demonstrate a deep-learning-based approach that can alleviate the defocus and motion blur of a microscopic image and output a sharper and cleaner image with retrieved fine details without prior knowledge of the blur type, blur extent and pathological stain. In this approach, a deep learning classifier is first trained to identify the image blur type. Then, two encoder-decoder networks are trained and used alone or in combination to deblur the input image. It is an end-to-end approach and introduces no corrugated artifacts as traditional blind deconvolution methods do. We test our approach on different types of pathology specimens and demonstrate great performance on image blur correction and the subsequent improvement on the diagnosis outcome of AI algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge