Dongqi Wang
TOP: Time Optimization Policy for Stable and Accurate Standing Manipulation with Humanoid Robots
Aug 01, 2025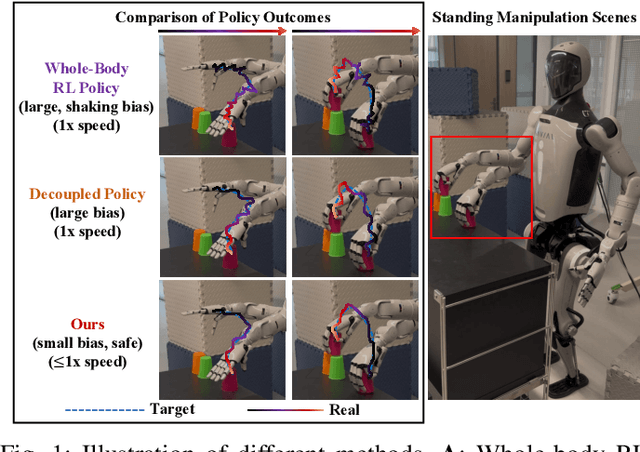
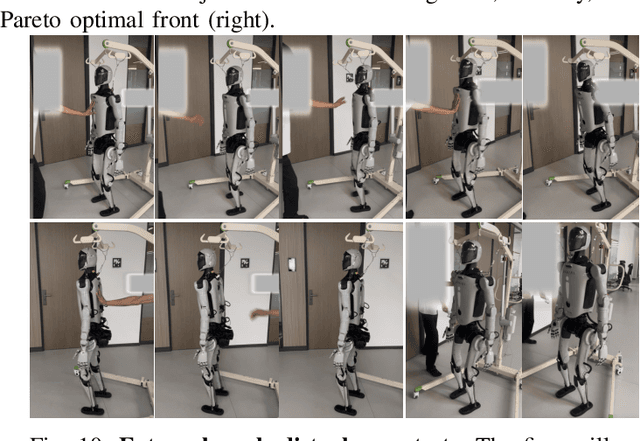

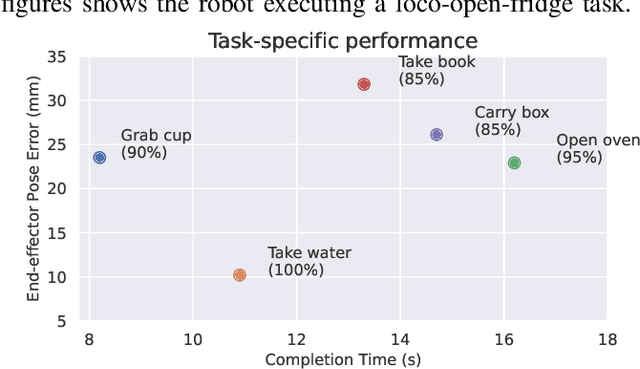
Abstract:Humanoid robots have the potential capability to perform a diverse range of manipulation tasks, but this is based on a robust and precise standing controller. Existing methods are either ill-suited to precisely control high-dimensional upper-body joints, or difficult to ensure both robustness and accuracy, especially when upper-body motions are fast. This paper proposes a novel time optimization policy (TOP), to train a standing manipulation control model that ensures balance, precision, and time efficiency simultaneously, with the idea of adjusting the time trajectory of upper-body motions but not only strengthening the disturbance resistance of the lower-body. Our approach consists of three parts. Firstly, we utilize motion prior to represent upper-body motions to enhance the coordination ability between the upper and lower-body by training a variational autoencoder (VAE). Then we decouple the whole-body control into an upper-body PD controller for precision and a lower-body RL controller to enhance robust stability. Finally, we train TOP method in conjunction with the decoupled controller and VAE to reduce the balance burden resulting from fast upper-body motions that would destabilize the robot and exceed the capabilities of the lower-body RL policy. The effectiveness of the proposed approach is evaluated via both simulation and real world experiments, which demonstrate the superiority on standing manipulation tasks stably and accurately. The project page can be found at https://anonymous.4open.science/w/top-258F/.
An eight-neuron network for quadruped locomotion with hip-knee joint control
Jun 19, 2024Abstract:The gait generator, which is capable of producing rhythmic signals for coordinating multiple joints, is an essential component in the quadruped robot locomotion control framework. The biological counterpart of the gait generator is the Central Pattern Generator (abbreviated as CPG), a small neural network consisting of interacting neurons. Inspired by this architecture, researchers have designed artificial neural networks composed of simulated neurons or oscillator equations. Despite the widespread application of these designed CPGs in various robot locomotion controls, some issues remain unaddressed, including: (1) Simplistic network designs often overlook the symmetry between signal and network structure, resulting in fewer gait patterns than those found in nature. (2) Due to minimal architectural consideration, quadruped control CPGs typically consist of only four neurons, which restricts the network's direct control to leg phases rather than joint coordination. (3) Gait changes are achieved by varying the neuron couplings or the assignment between neurons and legs, rather than through external stimulation. We apply symmetry theory to design an eight-neuron network, composed of Stein neuronal models, capable of achieving five gaits and coordinated control of the hip-knee joints. We validate the signal stability of this network as a gait generator through numerical simulations, which reveal various results and patterns encountered during gait transitions using neuronal stimulation. Based on these findings, we have developed several successful gait transition strategies through neuronal stimulations. Using a commercial quadruped robot model, we demonstrate the usability and feasibility of this network by implementing motion control and gait transitions.
Bridging the gap between target-based and cell-based drug discovery with a graph generative multi-task model
Aug 09, 2022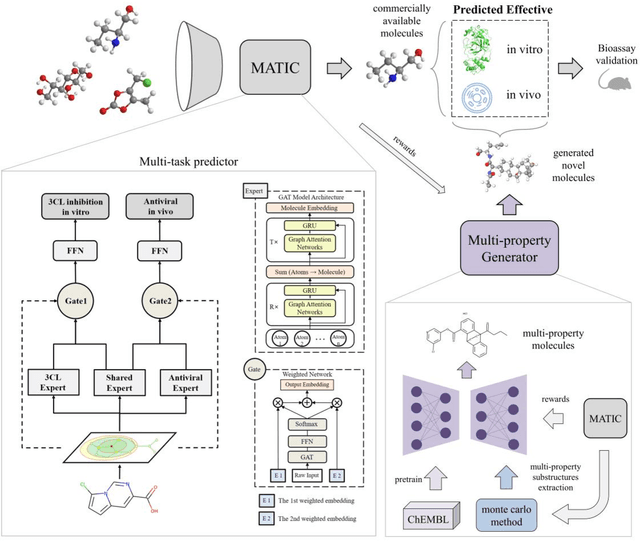
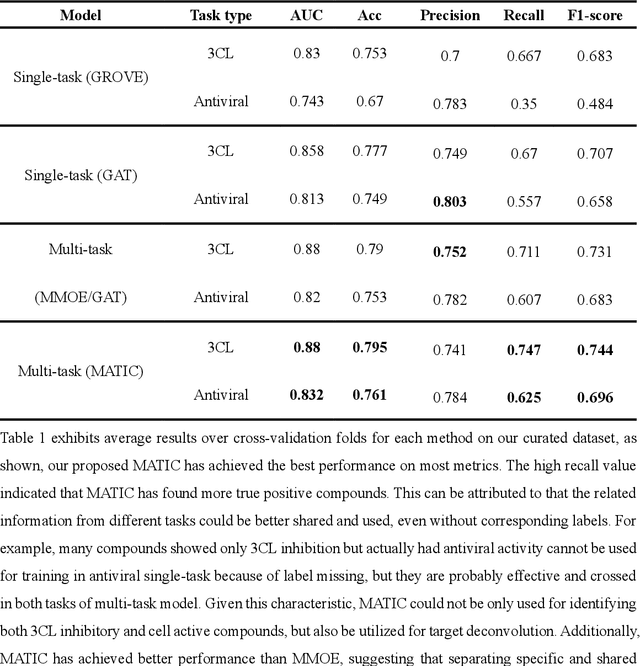
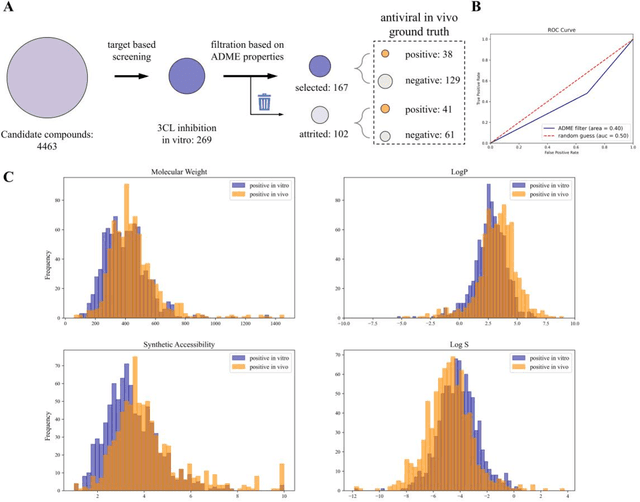
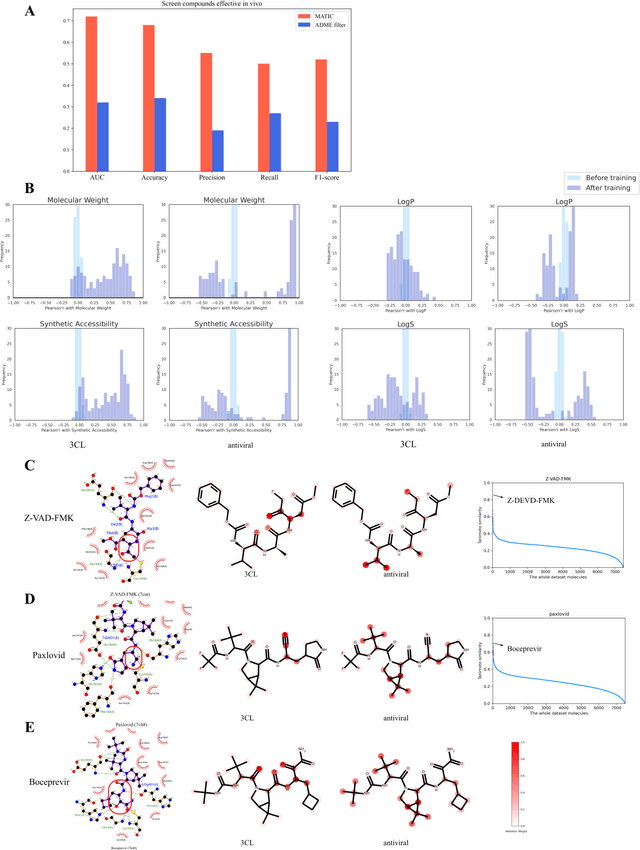
Abstract:Drug discovery is vitally important for protecting human against disease. Target-based screening is one of the most popular methods to develop new drugs in the past several decades. This method efficiently screens candidate drugs inhibiting target protein in vitro, but it often fails due to inadequate activity of the selected drugs in vivo. Accurate computational methods are needed to bridge this gap. Here, we propose a novel graph multi task deep learning model to identify compounds carrying both target inhibitory and cell active (MATIC) properties. On a carefully curated SARS-CoV-2 dataset, the proposed MATIC model shows advantages comparing with traditional method in screening effective compounds in vivo. Next, we explored the model interpretability and found that the learned features for target inhibition (in vitro) or cell active (in vivo) tasks are different with molecular property correlations and atom functional attentions. Based on these findings, we utilized a monte carlo based reinforcement learning generative model to generate novel multi-property compounds with both in vitro and in vivo efficacy, thus bridging the gap between target-based and cell-based drug discovery.
$\textit{latent}$-GLAT: Glancing at Latent Variables for Parallel Text Generation
Apr 05, 2022



Abstract:Recently, parallel text generation has received widespread attention due to its success in generation efficiency. Although many advanced techniques are proposed to improve its generation quality, they still need the help of an autoregressive model for training to overcome the one-to-many multi-modal phenomenon in the dataset, limiting their applications. In this paper, we propose $\textit{latent}$-GLAT, which employs the discrete latent variables to capture word categorical information and invoke an advanced curriculum learning technique, alleviating the multi-modality problem. Experiment results show that our method outperforms strong baselines without the help of an autoregressive model, which further broadens the application scenarios of the parallel decoding paradigm.
A Novel Architecture Slimming Method for Network Pruning and Knowledge Distillation
Feb 21, 2022



Abstract:Network pruning and knowledge distillation are two widely-known model compression methods that efficiently reduce computation cost and model size. A common problem in both pruning and distillation is to determine compressed architecture, i.e., the exact number of filters per layer and layer configuration, in order to preserve most of the original model capacity. In spite of the great advances in existing works, the determination of an excellent architecture still requires human interference or tremendous experimentations. In this paper, we propose an architecture slimming method that automates the layer configuration process. We start from the perspective that the capacity of the over-parameterized model can be largely preserved by finding the minimum number of filters preserving the maximum parameter variance per layer, resulting in a thin architecture. We formulate the determination of compressed architecture as a one-step orthogonal linear transformation, and integrate principle component analysis (PCA), where the variances of filters in the first several projections are maximized. We demonstrate the rationality of our analysis and the effectiveness of the proposed method through extensive experiments. In particular, we show that under the same overall compression rate, the compressed architecture determined by our method shows significant performance gain over baselines after pruning and distillation. Surprisingly, we find that the resulting layer-wise compression rates correspond to the layer sensitivities found by existing works through tremendous experimentations.
Non-Parametric Online Learning from Human Feedback for Neural Machine Translation
Sep 23, 2021



Abstract:We study the problem of online learning with human feedback in the human-in-the-loop machine translation, in which the human translators revise the machine-generated translations and then the corrected translations are used to improve the neural machine translation (NMT) system. However, previous methods require online model updating or additional translation memory networks to achieve high-quality performance, making them inflexible and inefficient in practice. In this paper, we propose a novel non-parametric online learning method without changing the model structure. This approach introduces two k-nearest-neighbor (KNN) modules: one module memorizes the human feedback, which is the correct sentences provided by human translators, while the other balances the usage of the history human feedback and original NMT models adaptively. Experiments conducted on EMEA and JRC-Acquis benchmarks demonstrate that our proposed method obtains substantial improvements on translation accuracy and achieves better adaptation performance with less repeating human correction operations.
A Novel Framework Integrating AI Model and Enzymological Experiments Promotes Identification of SARS-CoV-2 3CL Protease Inhibitors and Activity-based Probe
May 29, 2021
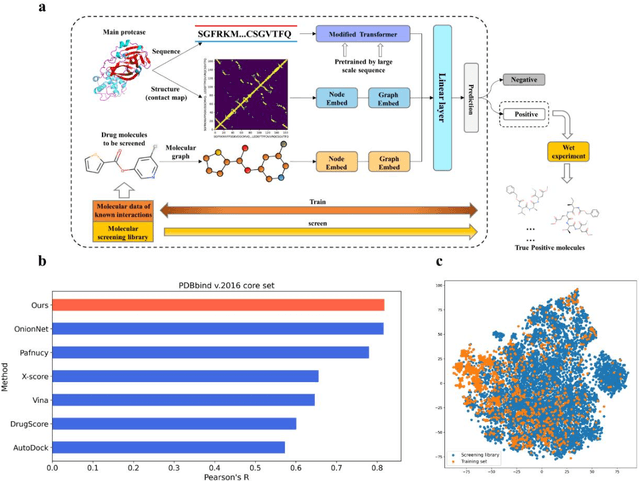


Abstract:The identification of protein-ligand interaction plays a key role in biochemical research and drug discovery. Although deep learning has recently shown great promise in discovering new drugs, there remains a gap between deep learning-based and experimental approaches. Here we propose a novel framework, named AIMEE, integrating AI Model and Enzymology Experiments, to identify inhibitors against 3CL protease of SARS-CoV-2, which has taken a significant toll on people across the globe. From a bioactive chemical library, we have conducted two rounds of experiments and identified six novel inhibitors with a hit rate of 29.41%, and four of them showed an IC50 value less than 3 {\mu}M. Moreover, we explored the interpretability of the central model in AIMEE, mapping the deep learning extracted features to domain knowledge of chemical properties. Based on this knowledge, a commercially available compound was selected and proven to be an activity-based probe of 3CLpro. This work highlights the great potential of combining deep learning models and biochemical experiments for intelligent iteration and expanding the boundaries of drug discovery.
Non-Autoregressive Translation by Learning Target Categorical Codes
Mar 21, 2021



Abstract:Non-autoregressive Transformer is a promising text generation model. However, current non-autoregressive models still fall behind their autoregressive counterparts in translation quality. We attribute this accuracy gap to the lack of dependency modeling among decoder inputs. In this paper, we propose CNAT, which learns implicitly categorical codes as latent variables into the non-autoregressive decoding. The interaction among these categorical codes remedies the missing dependencies and improves the model capacity. Experiment results show that our model achieves comparable or better performance in machine translation tasks, compared with several strong baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge