Huazhen Huang
Alt-MoE: Multimodal Alignment via Alternating Optimization of Multi-directional MoE with Unimodal Models
Sep 09, 2024



Abstract:Recent Large Multi-Modal Models (LMMs) have made significant advancements in multi-modal alignment by employing lightweight connection modules to facilitate the representation and fusion of knowledge from existing pre-trained uni-modal models. However, these methods still rely on modality-specific and direction-specific connectors, leading to compartmentalized knowledge representations and reduced computational efficiency, which limits the model's ability to form unified multi-modal representations. To address these issues, we introduce a novel training framework, Alt-MoE, which employs the Mixture of Experts (MoE) as a unified multi-directional connector across modalities, and employs a multi-step sequential alternating unidirectional alignment strategy, which converges to bidirectional alignment over iterations. The extensive empirical studies revealed the following key points: 1) Alt-MoE achieves competitive results by integrating diverse knowledge representations from uni-modal models. This approach seamlessly fuses the specialized expertise of existing high-performance uni-modal models, effectively synthesizing their domain-specific knowledge into a cohesive multi-modal representation. 2) Alt-MoE efficiently scales to new tasks and modalities without altering its model architecture or training strategy. Furthermore, Alt-MoE operates in latent space, supporting vector pre-storage and real-time retrieval via lightweight multi-directional MoE, thereby facilitating massive data processing. Our methodology has been validated on several well-performing uni-modal models (LLAMA3, Qwen2, and DINOv2), achieving competitive results on a wide range of downstream tasks and datasets.
Bridging the gap between target-based and cell-based drug discovery with a graph generative multi-task model
Aug 09, 2022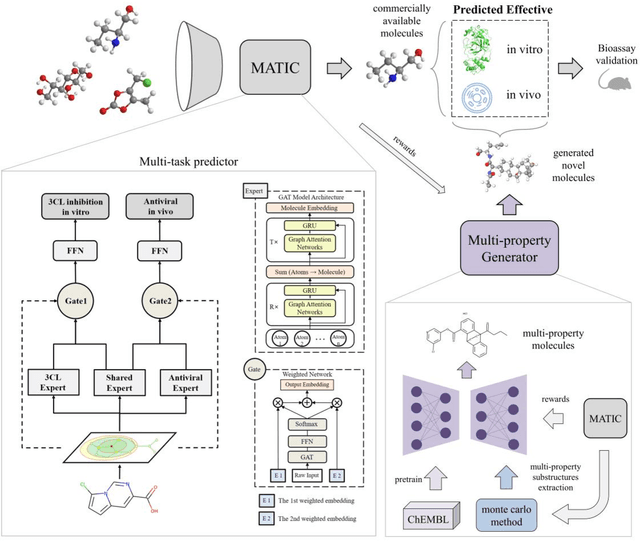
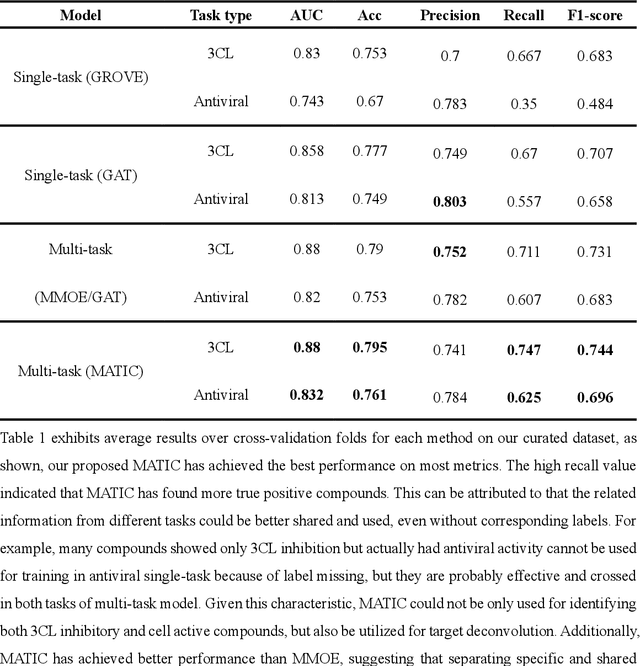
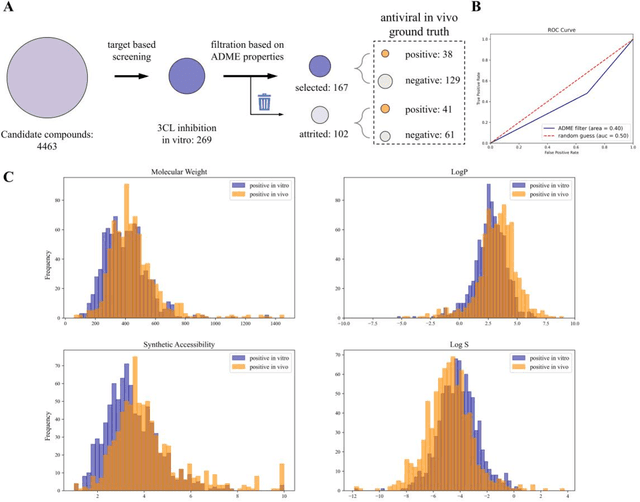
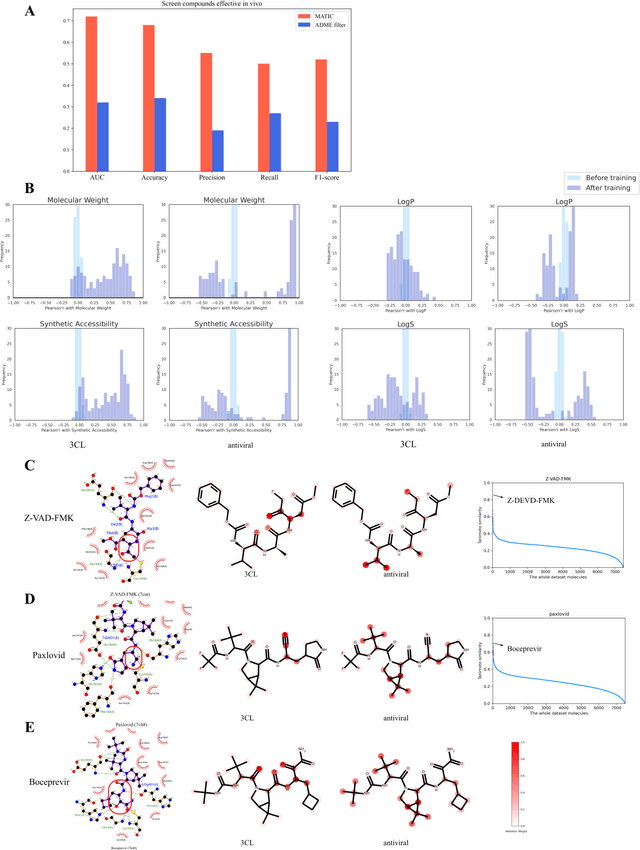
Abstract:Drug discovery is vitally important for protecting human against disease. Target-based screening is one of the most popular methods to develop new drugs in the past several decades. This method efficiently screens candidate drugs inhibiting target protein in vitro, but it often fails due to inadequate activity of the selected drugs in vivo. Accurate computational methods are needed to bridge this gap. Here, we propose a novel graph multi task deep learning model to identify compounds carrying both target inhibitory and cell active (MATIC) properties. On a carefully curated SARS-CoV-2 dataset, the proposed MATIC model shows advantages comparing with traditional method in screening effective compounds in vivo. Next, we explored the model interpretability and found that the learned features for target inhibition (in vitro) or cell active (in vivo) tasks are different with molecular property correlations and atom functional attentions. Based on these findings, we utilized a monte carlo based reinforcement learning generative model to generate novel multi-property compounds with both in vitro and in vivo efficacy, thus bridging the gap between target-based and cell-based drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge