Deniz Erdoğmuş
Active Learning For Repairable Hardware Systems With Partial Coverage
Mar 20, 2025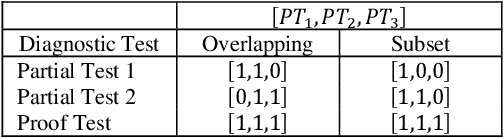
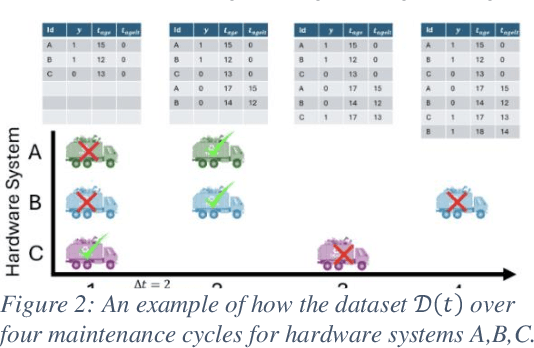
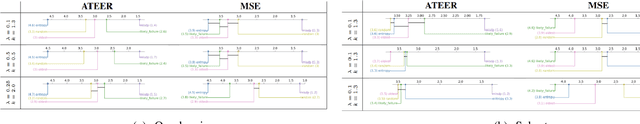
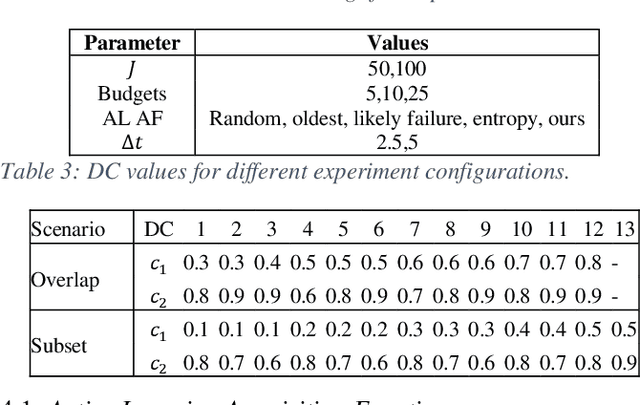
Abstract:Identifying the optimal diagnostic test and hardware system instance to infer reliability characteristics using field data is challenging, especially when constrained by fixed budgets and minimal maintenance cycles. Active Learning (AL) has shown promise for parameter inference with limited data and budget constraints in machine learning/deep learning tasks. However, AL for reliability model parameter inference remains underexplored for repairable hardware systems. It requires specialized AL Acquisition Functions (AFs) that consider hardware aging and the fact that a hardware system consists of multiple sub-systems, which may undergo only partial testing during a given diagnostic test. To address these challenges, we propose a relaxed Mixed Integer Semidefinite Program (MISDP) AL AF that incorporates Diagnostic Coverage (DC), Fisher Information Matrices (FIMs), and diagnostic testing budgets. Furthermore, we design empirical-based simulation experiments focusing on two diagnostic testing scenarios: (1) partial tests of a hardware system with overlapping subsystem coverage, and (2) partial tests where one diagnostic test fully subsumes the subsystem coverage of another. We evaluate our proposed approach against the most widely used AL AF in the literature (entropy), as well as several intuitive AL AFs tailored for reliability model parameter inference. Our proposed AF ranked best on average among the alternative AFs across 6,000 experimental configurations, with respect to Area Under the Curve (AUC) of the Absolute Total Expected Event Error (ATEER) and Mean Squared Error (MSE) curves, with statistical significance calculated at a 0.05 alpha level using a Friedman hypothesis test.
Learning Interpretable Deep Disentangled Neural Networks for Hyperspectral Unmixing
Oct 03, 2023



Abstract:Although considerable effort has been dedicated to improving the solution to the hyperspectral unmixing problem, non-idealities such as complex radiation scattering and endmember variability negatively impact the performance of most existing algorithms and can be very challenging to address. Recently, deep learning-based frameworks have been explored for hyperspectral umixing due to their flexibility and powerful representation capabilities. However, such techniques either do not address the non-idealities of the unmixing problem, or rely on black-box models which are not interpretable. In this paper, we propose a new interpretable deep learning method for hyperspectral unmixing that accounts for nonlinearity and endmember variability. The proposed method leverages a probabilistic variational deep-learning framework, where disentanglement learning is employed to properly separate the abundances and endmembers. The model is learned end-to-end using stochastic backpropagation, and trained using a self-supervised strategy which leverages benefits from semi-supervised learning techniques. Furthermore, the model is carefully designed to provide a high degree of interpretability. This includes modeling the abundances as a Dirichlet distribution, the endmembers using low-dimensional deep latent variable representations, and using two-stream neural networks composed of additive piecewise-linear/nonlinear components. Experimental results on synthetic and real datasets illustrate the performance of the proposed method compared to state-of-the-art algorithms.
Fetal-BET: Brain Extraction Tool for Fetal MRI
Oct 02, 2023



Abstract:Fetal brain extraction is a necessary first step in most computational fetal brain MRI pipelines. However, it has been a very challenging task due to non-standard fetal head pose, fetal movements during examination, and vastly heterogeneous appearance of the developing fetal brain and the neighboring fetal and maternal anatomy across various sequences and scanning conditions. Development of a machine learning method to effectively address this task requires a large and rich labeled dataset that has not been previously available. As a result, there is currently no method for accurate fetal brain extraction on various fetal MRI sequences. In this work, we first built a large annotated dataset of approximately 72,000 2D fetal brain MRI images. Our dataset covers the three common MRI sequences including T2-weighted, diffusion-weighted, and functional MRI acquired with different scanners. Moreover, it includes normal and pathological brains. Using this dataset, we developed and validated deep learning methods, by exploiting the power of the U-Net style architectures, the attention mechanism, multi-contrast feature learning, and data augmentation for fast, accurate, and generalizable automatic fetal brain extraction. Our approach leverages the rich information from multi-contrast (multi-sequence) fetal MRI data, enabling precise delineation of the fetal brain structures. Evaluations on independent test data show that our method achieves accurate brain extraction on heterogeneous test data acquired with different scanners, on pathological brains, and at various gestational stages. This robustness underscores the potential utility of our deep learning model for fetal brain imaging and image analysis.
A Multi-label Classification Approach to Increase Expressivity of EMG-based Gesture Recognition
Sep 13, 2023Abstract:Objective: The objective of the study is to efficiently increase the expressivity of surface electromyography-based (sEMG) gesture recognition systems. Approach: We use a problem transformation approach, in which actions were subset into two biomechanically independent components - a set of wrist directions and a set of finger modifiers. To maintain fast calibration time, we train models for each component using only individual gestures, and extrapolate to the full product space of combination gestures by generating synthetic data. We collected a supervised dataset with high-confidence ground truth labels in which subjects performed combination gestures while holding a joystick, and conducted experiments to analyze the impact of model architectures, classifier algorithms, and synthetic data generation strategies on the performance of the proposed approach. Main Results: We found that a problem transformation approach using a parallel model architecture in combination with a non-linear classifier, along with restricted synthetic data generation, shows promise in increasing the expressivity of sEMG-based gestures with a short calibration time. Significance: sEMG-based gesture recognition has applications in human-computer interaction, virtual reality, and the control of robotic and prosthetic devices. Existing approaches require exhaustive model calibration. The proposed approach increases expressivity without requiring users to demonstrate all combination gesture classes. Our results may be extended to larger gesture vocabularies and more complicated model architectures.
A Vision for Cleaner Rivers: Harnessing Snapshot Hyperspectral Imaging to Detect Macro-Plastic Litter
Jul 22, 2023



Abstract:Plastic waste entering the riverine harms local ecosystems leading to negative ecological and economic impacts. Large parcels of plastic waste are transported from inland to oceans leading to a global scale problem of floating debris fields. In this context, efficient and automatized monitoring of mismanaged plastic waste is paramount. To address this problem, we analyze the feasibility of macro-plastic litter detection using computational imaging approaches in river-like scenarios. We enable near-real-time tracking of partially submerged plastics by using snapshot Visible-Shortwave Infrared hyperspectral imaging. Our experiments indicate that imaging strategies associated with machine learning classification approaches can lead to high detection accuracy even in challenging scenarios, especially when leveraging hyperspectral data and nonlinear classifiers. All code, data, and models are available online: https://github.com/RIVeR-Lab/hyperspectral_macro_plastic_detection.
Hybrid Neural Network Augmented Physics-based Models for Nonlinear Filtering
Apr 13, 2022



Abstract:In this paper we present a hybrid neural network augmented physics-based modeling (APBM) framework for Bayesian nonlinear latent space estimation. The proposed APBM strategy allows for model adaptation when new operation conditions come into play or the physics-based model is insufficient (or incomplete) to properly describe the latent phenomenon. One advantage of the APBMs and our estimation procedure is the capability of maintaining the physical interpretability of estimated states. Furthermore, we propose a constraint filtering approach to control the neural network contributions to the overall model. We also exploit assumed density filtering techniques and cubature integration rules to present a flexible estimation strategy that can easily deal with nonlinear models and high-dimensional latent spaces. Finally, we demonstrate the efficacy of our methodology by leveraging a target tracking scenario with nonlinear and incomplete measurement and acceleration models, respectively.
Equivariant Deep Dynamical Model for Motion Prediction
Nov 02, 2021



Abstract:Learning representations through deep generative modeling is a powerful approach for dynamical modeling to discover the most simplified and compressed underlying description of the data, to then use it for other tasks such as prediction. Most learning tasks have intrinsic symmetries, i.e., the input transformations leave the output unchanged, or the output undergoes a similar transformation. The learning process is, however, usually uninformed of these symmetries. Therefore, the learned representations for individually transformed inputs may not be meaningfully related. In this paper, we propose an SO(3) equivariant deep dynamical model (EqDDM) for motion prediction that learns a structured representation of the input space in the sense that the embedding varies with symmetry transformations. EqDDM is equipped with equivariant networks to parameterize the state-space emission and transition models. We demonstrate the superior predictive performance of the proposed model on various motion data.
Model-Based Deep Autoencoder Networks for Nonlinear Hyperspectral Unmixing
Apr 17, 2021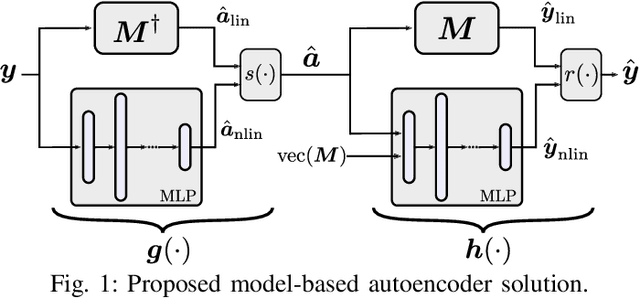
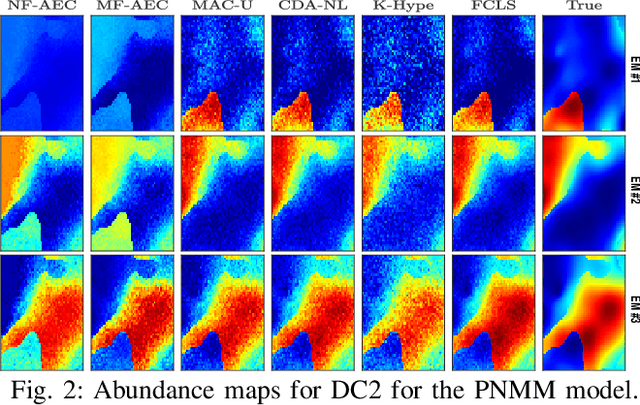
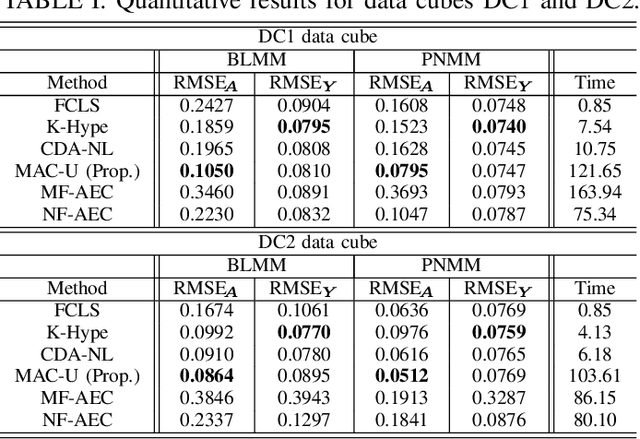
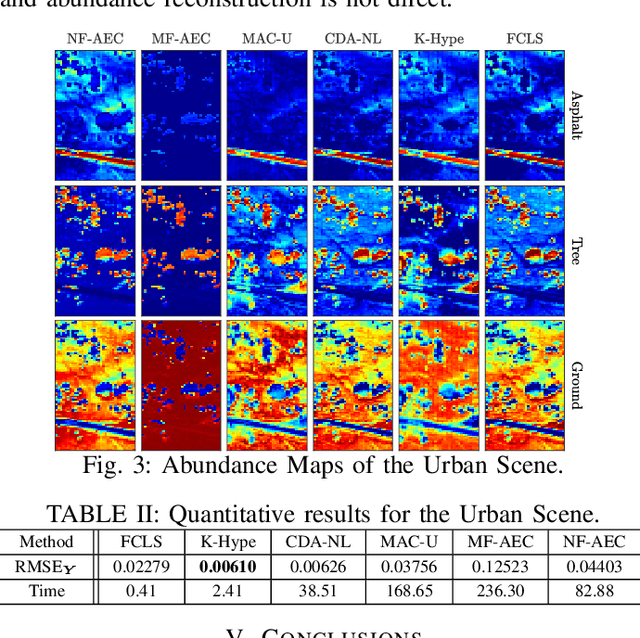
Abstract:Autoencoder (AEC) networks have recently emerged as a promising approach to perform unsupervised hyperspectral unmixing (HU) by associating the latent representations with the abundances, the decoder with the mixing model and the encoder with its inverse. AECs are especially appealing for nonlinear HU since they lead to unsupervised and model-free algorithms. However, existing approaches fail to explore the fact that the encoder should invert the mixing process, which might reduce their robustness. In this paper, we propose a model-based AEC for nonlinear HU by considering the mixing model a nonlinear fluctuation over a linear mixture. Differently from previous works, we show that this restriction naturally imposes a particular structure to both the encoder and to the decoder networks. This introduces prior information in the AEC without reducing the flexibility of the mixing model. Simulations with synthetic and real data indicate that the proposed strategy improves nonlinear HU.
Prediction of Epilepsy Development in Traumatic Brain Injury Patients from Diffusion Weighted MRI
May 01, 2020

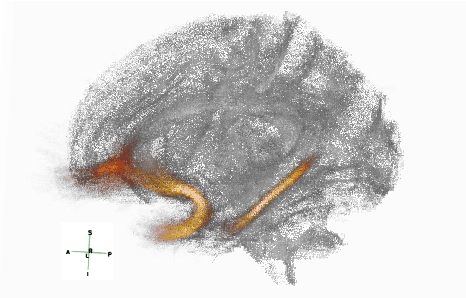
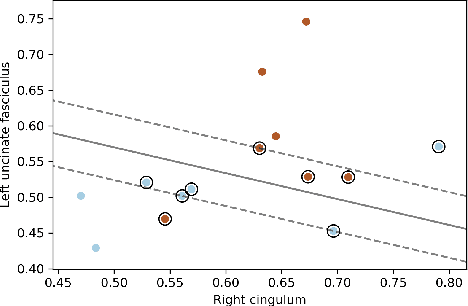
Abstract:Post-traumatic epilepsy (PTE) is a life-long complication of traumatic brain injury (TBI) and is a major public health problem that has an estimated incidence that ranges from 2%-50%, depending on the severity of the TBI. Currently, the pathomechanism that in-duces epileptogenesis in TBI patients is unclear, and one of the most challenging goals in the epilepsy community is to predict which TBI patients will develop epilepsy. In this work, we used diffusion-weighted imaging (DWI) of 14 TBI patients recruited in the Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx)to measure and analyze fractional anisotropy (FA), obtained from tract-based spatial statistic (TBSS) analysis. Then we used these measurements to train two support vector machine (SVM) models to predict which TBI patients have developed epilepsy. Our approach, tested on these 14 patients with a leave-two-out cross-validation, allowed us to obtain an accuracy of 0.857 $\pm$ 0.18 (with a 95% level of confidence), demonstrating it to be potentially promising for the early characterization of PTE.
Mapping Motor Cortex Stimulation to Muscle Responses: A Deep Neural Network Modeling Approach
Feb 14, 2020



Abstract:A deep neural network (DNN) that can reliably model muscle responses from corresponding brain stimulation has the potential to increase knowledge of coordinated motor control for numerous basic science and applied use cases. Such cases include the understanding of abnormal movement patterns due to neurological injury from stroke, and stimulation based interventions for neurological recovery such as paired associative stimulation. In this work, potential DNN models are explored and the one with the minimum squared errors is recommended for the optimal performance of the M2M-Net, a network that maps transcranial magnetic stimulation of the motor cortex to corresponding muscle responses, using: a finite element simulation, an empirical neural response profile, a convolutional autoencoder, a separate deep network mapper, and recordings of multi-muscle activation. We discuss the rationale behind the different modeling approaches and architectures, and contrast their results. Additionally, to obtain a comparative insight of the trade-off between complexity and performance analysis, we explore different techniques, including the extension of two classical information criteria for M2M-Net. Finally, we find that the model analogous to mapping the motor cortex stimulation to a combination of direct and synergistic connection to the muscles performs the best, when the neural response profile is used at the input.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge