Razieh Faghihpirayesh
UniMo: Universal Motion Correction For Medical Images without Network Retraining
Sep 21, 2024



Abstract:In this paper, we introduce a Universal Motion Correction (UniMo) framework, leveraging deep neural networks to tackle the challenges of motion correction across diverse imaging modalities. Our approach employs advanced neural network architectures with equivariant filters, overcoming the limitations of current models that require iterative inference or retraining for new image modalities. UniMo enables one-time training on a single modality while maintaining high stability and adaptability for inference across multiple unseen image modalities. We developed a joint learning framework that integrates multimodal knowledge from both shape and images that faithfully improve motion correction accuracy despite image appearance variations. UniMo features a geometric deformation augmenter that enhances the robustness of global motion correction by addressing any local deformations whether they are caused by object deformations or geometric distortions, and also generates augmented data to improve the training process. Our experimental results, conducted on various datasets with four different image modalities, demonstrate that UniMo surpasses existing motion correction methods in terms of accuracy. By offering a comprehensive solution to motion correction, UniMo marks a significant advancement in medical imaging, especially in challenging applications with wide ranges of motion, such as fetal imaging. The code for this work is available online, https://github.com/IntelligentImaging/UNIMO/.
SpaER: Learning Spatio-temporal Equivariant Representations for Fetal Brain Motion Tracking
Jul 29, 2024


Abstract:In this paper, we introduce SpaER, a pioneering method for fetal motion tracking that leverages equivariant filters and self-attention mechanisms to effectively learn spatio-temporal representations. Different from conventional approaches that statically estimate fetal brain motions from pairs of images, our method dynamically tracks the rigid movement patterns of the fetal head across temporal and spatial dimensions. Specifically, we first develop an equivariant neural network that efficiently learns rigid motion sequences through low-dimensional spatial representations of images. Subsequently, we learn spatio-temporal representations by incorporating time encoding and self-attention neural network layers. This approach allows for the capture of long-term dependencies of fetal brain motion and addresses alignment errors due to contrast changes and severe motion artifacts. Our model also provides a geometric deformation estimation that properly addresses image distortions among all time frames. To the best of our knowledge, our approach is the first to learn spatial-temporal representations via deep neural networks for fetal motion tracking without data augmentation. We validated our model using real fetal echo-planar images with simulated and real motions. Our method carries significant potential value in accurately measuring, tracking, and correcting fetal motion in fetal MRI sequences.
Fetal-BET: Brain Extraction Tool for Fetal MRI
Oct 02, 2023



Abstract:Fetal brain extraction is a necessary first step in most computational fetal brain MRI pipelines. However, it has been a very challenging task due to non-standard fetal head pose, fetal movements during examination, and vastly heterogeneous appearance of the developing fetal brain and the neighboring fetal and maternal anatomy across various sequences and scanning conditions. Development of a machine learning method to effectively address this task requires a large and rich labeled dataset that has not been previously available. As a result, there is currently no method for accurate fetal brain extraction on various fetal MRI sequences. In this work, we first built a large annotated dataset of approximately 72,000 2D fetal brain MRI images. Our dataset covers the three common MRI sequences including T2-weighted, diffusion-weighted, and functional MRI acquired with different scanners. Moreover, it includes normal and pathological brains. Using this dataset, we developed and validated deep learning methods, by exploiting the power of the U-Net style architectures, the attention mechanism, multi-contrast feature learning, and data augmentation for fast, accurate, and generalizable automatic fetal brain extraction. Our approach leverages the rich information from multi-contrast (multi-sequence) fetal MRI data, enabling precise delineation of the fetal brain structures. Evaluations on independent test data show that our method achieves accurate brain extraction on heterogeneous test data acquired with different scanners, on pathological brains, and at various gestational stages. This robustness underscores the potential utility of our deep learning model for fetal brain imaging and image analysis.
Deep Learning Framework for Real-time Fetal Brain Segmentation in MRI
May 02, 2022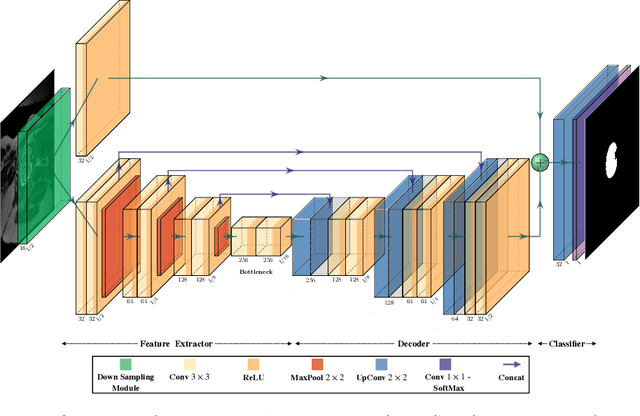


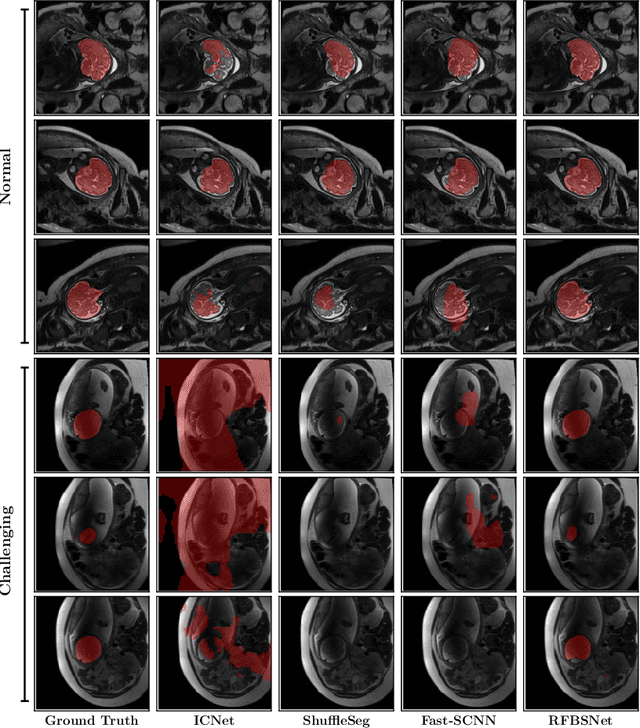
Abstract:Fetal brain segmentation is an important first step for slice-level motion correction and slice-to-volume reconstruction in fetal MRI. Fast and accurate segmentation of the fetal brain on fetal MRI is required to achieve real-time fetal head pose estimation and motion tracking for slice re-acquisition and steering. To address this critical unmet need, in this work we analyzed the speed-accuracy performance of a variety of deep neural network models, and devised a symbolically small convolutional neural network that combines spatial details at high resolution with context features extracted at lower resolutions. We used multiple branches with skip connections to maintain high accuracy while devising a parallel combination of convolution and pooling operations as an input downsampling module to further reduce inference time. We trained our model as well as eight alternative, state-of-the-art networks with manually-labeled fetal brain MRI slices and tested on two sets of normal and challenging test cases. Experimental results show that our network achieved the highest accuracy and lowest inference time among all of the compared state-of-the-art real-time segmentation methods. We achieved average Dice scores of 97.99\% and 84.04\% on the normal and challenging test sets, respectively, with an inference time of 3.36 milliseconds per image on an NVIDIA GeForce RTX 2080 Ti. Code, data, and the trained models are available at https://github.com/bchimagine/real_time_fetal_brain_segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge